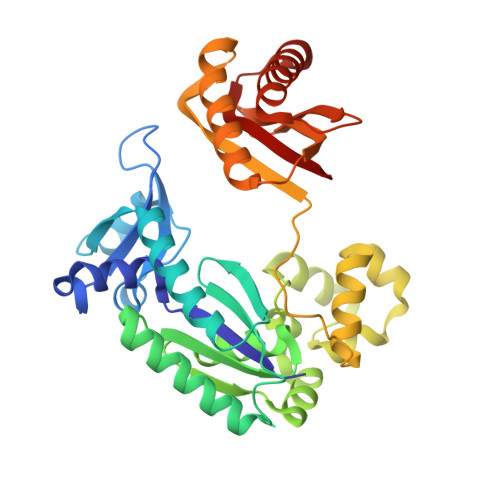
Structural mechanism of replication stalling on a bulky amino-polycyclic aromatic hydrocarbon DNA adduct by a y family DNA polymerase.
Kirouac, K.N., Basu, A.K., Ling, H.(2013) J Mol Biology 425: 4167-4176
- PubMed: 23876706
- DOI: https://doi.org/10.1016/j.jmb.2013.07.020
- Primary Citation of Related Structures:
4FBT, 4FBU - PubMed Abstract:
Polycyclic aromatic hydrocarbons and their nitro derivatives are culprits of the detrimental health effects of environmental pollution. These hydrophobic compounds metabolize to reactive species and attach to DNA producing bulky lesions, such as N-[deoxyguanosine-8-yl]-1-aminopyrene (APG), in genomic DNA. The bulky adducts block DNA replication by high-fidelity polymerases and compromise replication fidelities and efficiencies by specialized lesion bypass polymerases. Here we present three crystal structures of the DNA polymerase Dpo4, a model translesion DNA polymerase of the Y family, in complex with APG-lesion-containing DNA in pre-insertion and extension stages. APG is captured in two conformations in the pre-insertion complex; one is highly exposed to the solvent, whereas the other is harbored in a shallow cleft between the finger and unique Y family little finger domain. In contrast, APG is in a single conformation at the extension stage, in which the pyrene ring is sandwiched between the little finger domain and a base from the turning back single-stranded template strand. Strikingly, a nucleotide intercalates the DNA helix to form a quaternary complex with Dpo4, DNA, and an incoming nucleotide, which stabilizes the distorted DNA structure at the extension stage. The unique APG DNA conformations in Dpo4 inhibit DNA translocation through the polymerase active site for APG bypass. We also modeled an insertion complex that illustrates a solvent-exposed pyrene ring contributing to an unstable insertion state. The structural work combined with our lesion replication assays provides a novel structural mechanism on bypass of DNA adducts containing polycyclic aromatic hydrocarbon moieties.
- Department of Biochemistry, University of Western Ontario, London, Ontario, Canada N6A 5C1.
Organizational Affiliation: