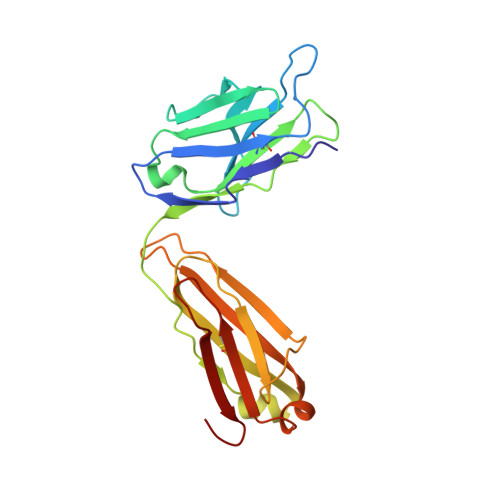
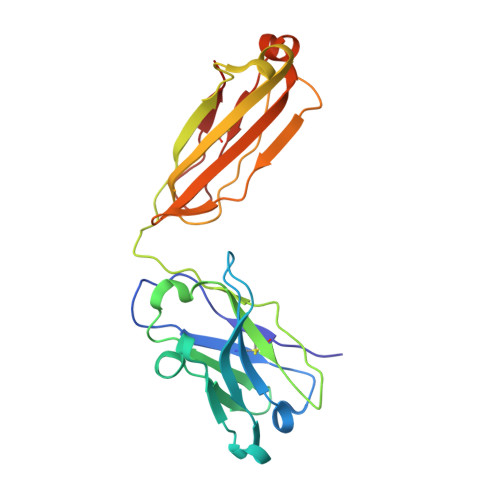
Three-dimensional structure of a fluorescein-Fab complex crystallized in 2-methyl-2,4-pentanediol.
Herron, J.N., He, X.M., Mason, M.L., Voss Jr., E.W., Edmundson, A.B.(1989) Proteins 5: 271-280
- PubMed: 2508085
- DOI: https://doi.org/10.1002/prot.340050404
- Primary Citation of Related Structures:
4FAB - PubMed Abstract:
The crystal structure of a fluorescein-Fab (4-4-20) complex was determined at 2.7 A resolution by molecular replacement methods. The starting model was the refined 2.7 A structure of unliganded Fab from an autoantibody (BV04-01) with specificity for single-stranded DNA. In the 4-4-20 complex fluorescein fits tightly into a relatively deep slot formed by a network of tryptophan and tyrosine side chains. The planar xanthonyl ring of the hapten is accommodated at the bottom of the slot while the phenylcarboxyl group interfaces with solvent. Tyrosine 37 (light chain) and tryptophan 33 (heavy chain) flank the xanthonyl group and tryptophan 101 (light chain) provides the floor of the combining site. Tyrosine 103 (heavy chain) is situated near the phenyl ring of the hapten and tyrosine 102 (heavy chain) forms part of the boundary of the slot. Histidine 31 and arginine 39 of the light chain are located in positions adjacent to the two enolic groups at opposite ends of the xanthonyl ring, and thus account for neutralization of one of two negative charges in the haptenic dianion. Formation of an enol-arginine ion pair in a region of low dielectric constant may account for an incremental increase in affinity of 2-3 orders of magnitude in the 4-4-20 molecule relative to other members of an idiotypic family of monoclonal antifluorescyl antibodies. The phenyl carboxyl group of fluorescein appears to be hydrogen bonded to the phenolic hydroxyl group of tyrosine 37 of the light chain. A molecule of 2-methyl-2,4-pentanediol (MPD), trapped in the interface of the variable domains just below the fluorescein binding site, may be partly responsible for the decrease in affinity for the hapten in MPD.
- Department of Biology, University of Utah, Salt Lake City 84112.
Organizational Affiliation: