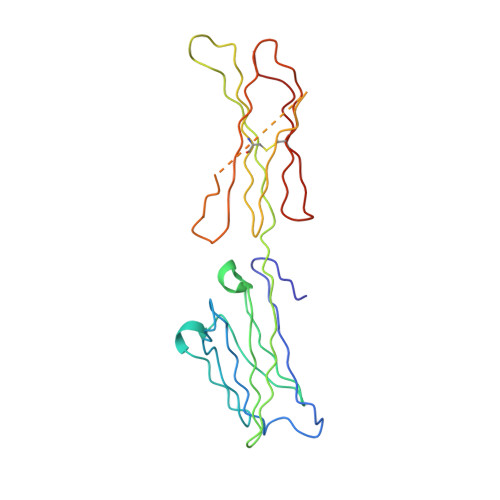
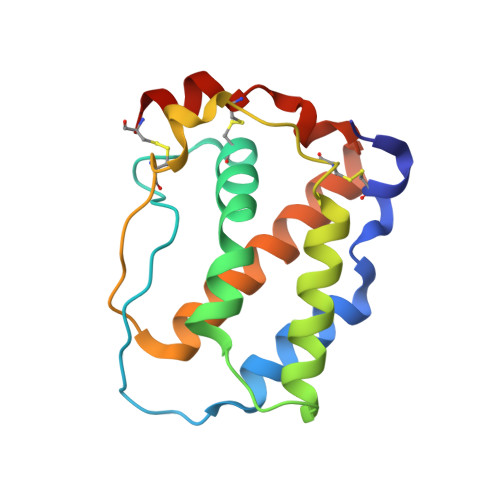
Multipronged attenuation of macrophage-colony stimulating factor signaling by Epstein-Barr virus BARF1.
Shim, A.H., Chang, R.A., Chen, X., Longnecker, R., He, X.(2012) Proc Natl Acad Sci U S A 109: 12962-12967
- PubMed: 22826234
- DOI: https://doi.org/10.1073/pnas.1205309109
- Primary Citation of Related Structures:
4FA8 - PubMed Abstract:
The ubiquitous EBV causes infectious mononucleosis and is associated with several types of cancers. The EBV genome encodes an early gene product, BARF1, which contributes to pathogenesis, potentially through growth-altering and immune-modulating activities, but the mechanisms for such activities are poorly understood. We have determined the crystal structure of BARF1 in complex with human macrophage-colony stimulating factor (M-CSF), a hematopoietic cytokine with pleiotropic functions in development and immune response. BARF1 and M-CSF form a high-affinity, stable, ring-like complex in both solution and the crystal, with a BARF1 hexameric ring surrounded by three M-CSF dimers in triangular array. The binding of BARF1 to M-CSF dramatically reduces but does not completely abolish M-CSF binding and signaling through its cognate receptor FMS. A three-pronged down-regulation mechanism is proposed to explain the biological effect of BARF1 on M-CSF:FMS signaling. These prongs entail control of the circulating and effective local M-CSF concentration, perturbation of the receptor-binding surface of M-CSF, and imposition of an unfavorable global orientation of the M-CSF dimer. Each prong may reduce M-CSF:FMS signaling to a limited extent but in combination may alter M-CSF:FMS signaling dramatically. The downregulating mechanism of BARF1 underlines a viral modulation strategy, and provides a basis for understanding EBV pathogenesis.
- Department of Molecular Pharmacology and Biological Chemistry, Feinberg School of Medicine, Northwestern University, Chicago, IL 60611, USA.
Organizational Affiliation: