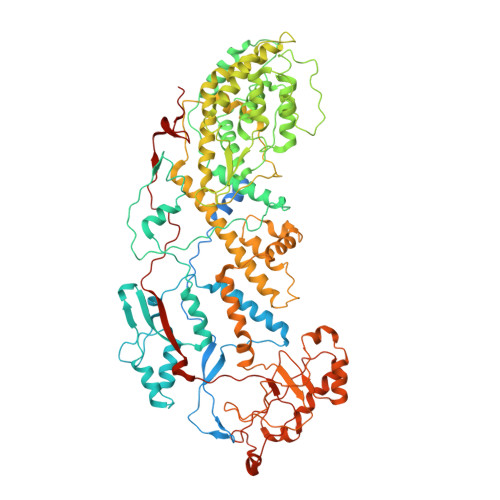
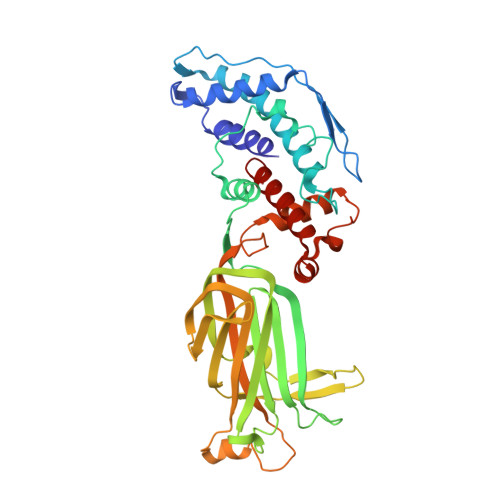
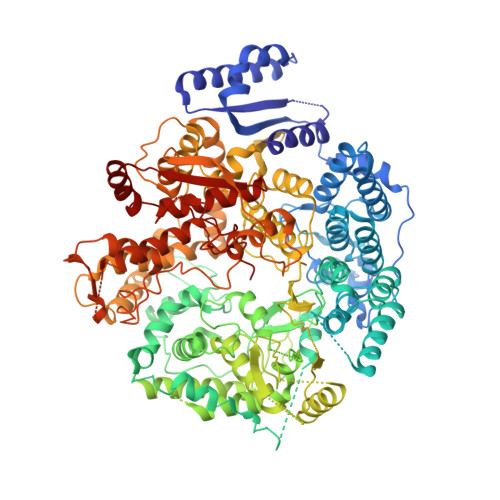
Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
Estrozi, L.F., Settembre, E.C., Goret, G., McClain, B., Zhang, X., Chen, J.Z., Grigorieff, N., Harrison, S.C.(2013) J Mol Biology 425: 124-132
- PubMed: 23089332
- DOI: https://doi.org/10.1016/j.jmb.2012.10.011
- Primary Citation of Related Structures:
4AU6, 4F5X - PubMed Abstract:
Double-stranded RNA (dsRNA) viruses transcribe and replicate RNA within an assembled, inner capsid particle; only plus-sense mRNA emerges into the intracellular milieu. During infectious entry of a rotavirus particle, the outer layer of its three-layer structure dissociates, delivering the inner double-layered particle (DLP) into the cytosol. DLP structures determined by X-ray crystallography and electron cryomicroscopy (cryoEM) show that the RNA coils uniformly into the particle interior, avoiding a "fivefold hub" of more structured density projecting inward from the VP2 shell of the DLP along each of the twelve 5-fold axes. Analysis of the X-ray crystallographic electron density map suggested that principal contributors to the hub are the N-terminal arms of VP2, but reexamination of the cryoEM map has shown that many features come from a molecule of VP1, randomly occupying five equivalent and partly overlapping positions. We confirm here that the electron density in the X-ray map leads to the same conclusion, and we describe the functional implications of the orientation and position of the polymerase. The exit channel for the nascent transcript directs the nascent transcript toward an opening along the 5-fold axis. The template strand enters from within the particle, and the dsRNA product of the initial replication step exits in a direction tangential to the inner surface of the VP2 shell, allowing it to coil optimally within the DLP. The polymerases of reoviruses appear to have similar positions and functional orientations.
- European Molecular Biology Laboratory, Grenoble Outstation, 6 Rue Jules Horowitz, Grenoble 38042, France. estrozi@embl.fr
Organizational Affiliation: