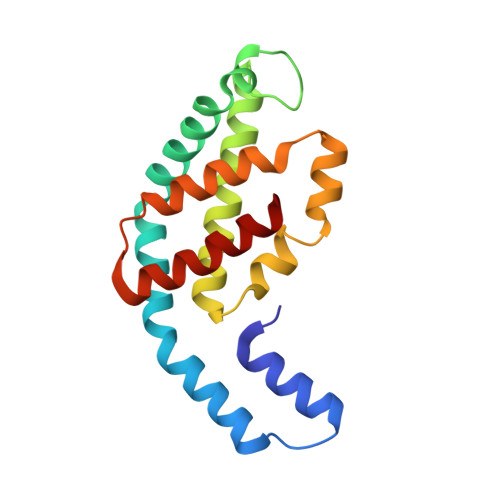
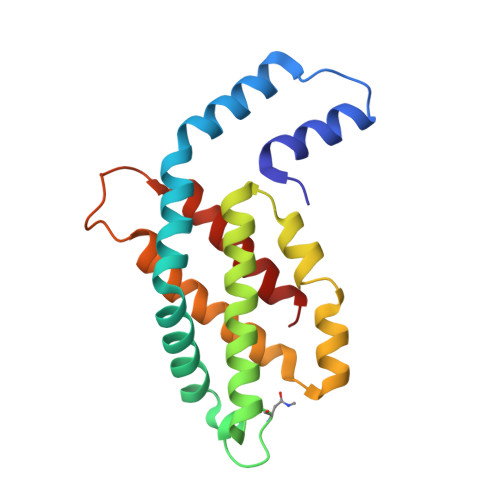
Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly.
Marx, A., Adir, N.(2013) Biochim Biophys Acta 1827: 311-318
- PubMed: 23201474
- DOI: https://doi.org/10.1016/j.bbabio.2012.11.006
- Primary Citation of Related Structures:
4F0T, 4F0U, 4GXE, 4GY3, 4H0M - PubMed Abstract:
X-ray crystal structures of the isolated phycobiliprotein components of the phycobilisome have provided high resolution details to the description of this light harvesting complex at different levels of complexity and detail. The linker-independent assembly of trimers into hexamers in crystal lattices of previously determined structures has been observed in almost all of the phycocyanin (PC) and allophycocyanin (APC) structures available in the Protein Data Bank. In this paper we describe the X-ray crystal structures of PC and APC from Synechococcus elongatus sp. PCC 7942, PC from Synechocystis sp. PCC 6803 and PC from Thermosynechococcus vulcanus crystallized in the presence of urea. All five structures are highly similar to other PC and APC structures on the levels of subunits, monomers and trimers. The Synechococcus APC forms a unique loose hexamer that may show the structural requirements for core assembly and rod attachment. While the Synechococcus PC assembles into the canonical hexamer, it does not further assemble into rods. Unlike most PC structures, the Synechocystis PC fails to form hexamers. Addition of low concentrations of urea to T. vulcanus PC inhibits this proteins propensity to form hexamers, resulting in a crystal lattice composed of trimers. The molecular source of these differences in assembly and their relevance to the phycobilisome structure is discussed.
- Schulich Faculty of Chemistry, Technion-Israel Institute of Technology, Haifa, Israel.
Organizational Affiliation: