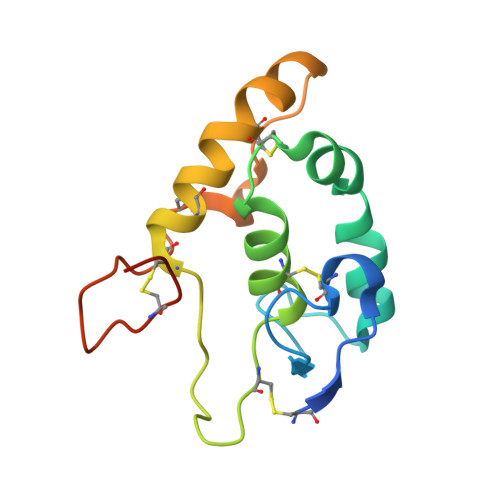
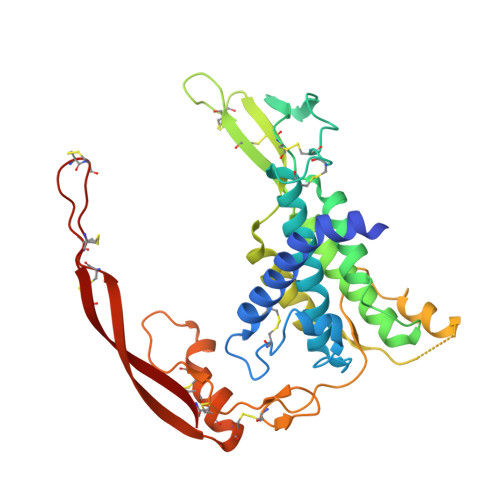
Structural basis of Wnt recognition by Frizzled.
Janda, C.Y., Waghray, D., Levin, A.M., Thomas, C., Garcia, K.C.(2012) Science 337: 59-64
- PubMed: 22653731
- DOI: https://doi.org/10.1126/science.1222879
- Primary Citation of Related Structures:
4F0A - PubMed Abstract:
Wnts are lipid-modified morphogens that play critical roles in development principally through engagement of Frizzled receptors. The 3.25 angstrom structure of Xenopus Wnt8 (XWnt8) in complex with mouse Frizzled-8 (Fz8) cysteine-rich domain (CRD) reveals an unusual two-domain Wnt structure, not obviously related to known protein folds, resembling a "hand" with "thumb" and "index" fingers extended to grasp the Fz8-CRD at two distinct binding sites. One site is dominated by a palmitoleic acid lipid group projecting from serine 187 at the tip of Wnt's thumb into a deep groove in the Fz8-CRD. In the second binding site, the conserved tip of Wnt's "index finger" forms hydrophobic amino acid contacts with a depression on the opposite side of the Fz8-CRD. The conservation of amino acids in both interfaces appears to facilitate ligand-receptor cross-reactivity, which has important implications for understanding Wnt's functional pleiotropy and for developing Wnt-based drugs for cancer and regenerative medicine.
- Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: