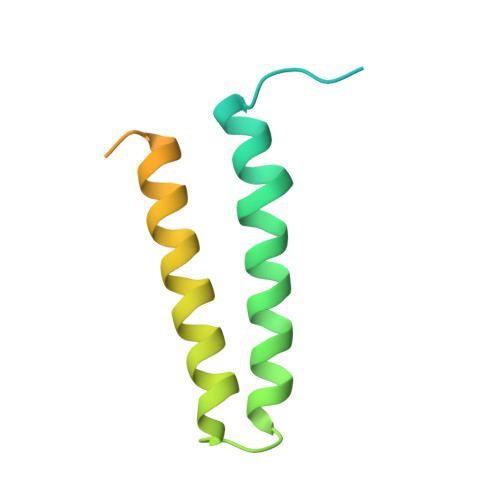
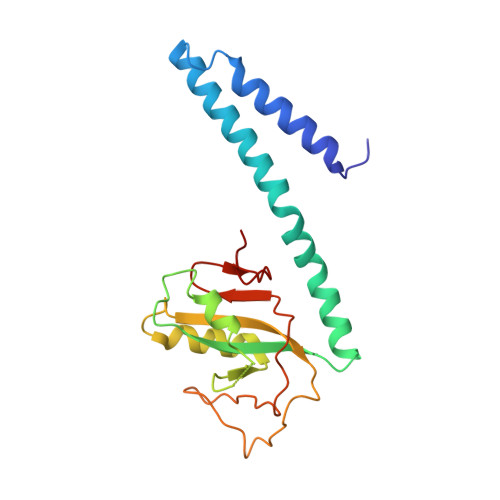
IcmQ in the Type 4b secretion system contains an NAD+ binding domain.
Farelli, J.D., Gumbart, J.C., Akey, I.V., Hempstead, A., Amyot, W., Head, J.F., McKnight, C.J., Isberg, R.R., Akey, C.W.(2013) Structure 21: 1361-1373
- PubMed: 23850453
- DOI: https://doi.org/10.1016/j.str.2013.05.017
- Primary Citation of Related Structures:
4EYY - PubMed Abstract:
A Type 4b secretion system (T4bSS) is required for Legionella growth in alveolar macrophages. IcmQ associates with IcmR, binds to membranes, and has a critical role in the T4bSS. We have now solved a crystal structure of IcmR-IcmQ to further our understanding of this complex. This structure revealed an amphipathic four-helix bundle, formed by IcmR and the N-terminal domain of IcmQ, which is linked to a novel C-terminal domain of IcmQ (Qc) by a linker helix. The Qc domain has structural homology with ADP ribosyltransferase domains in certain bacterial toxins and binds NAD(+) with a dissociation constant in the physiological range. Structural homology and molecular dynamics were used to identify an extended NAD(+) binding site on Qc, and the resulting model was tested by mutagenesis and binding assays. Based on the data, we suggest that IcmR-IcmQ binds to membranes, where it may interact with, or perhaps modify, a protein in the T4bSS when NAD(+) is bound.
- Department of Physiology and Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118-2526, USA.
Organizational Affiliation: