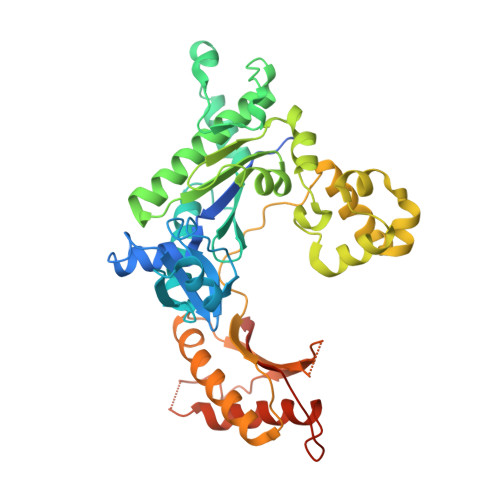
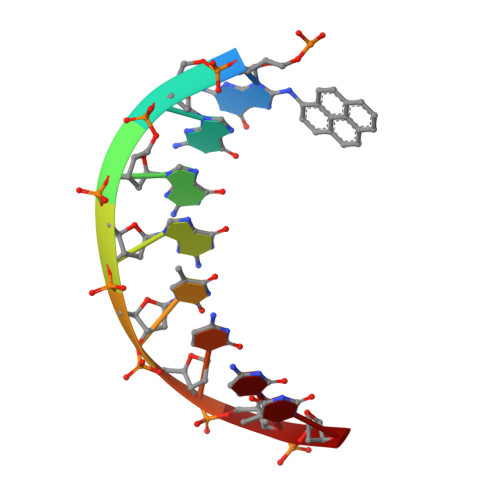
Replication of a carcinogenic nitropyrene DNA lesion by human Y-family DNA polymerase.
Kirouac, K.N., Basu, A.K., Ling, H.(2013) Nucleic Acids Res 41: 2060-2071
- PubMed: 23268450
- DOI: https://doi.org/10.1093/nar/gks1296
- Primary Citation of Related Structures:
4EYH, 4EYI - PubMed Abstract:
Nitrated polycyclic aromatic hydrocarbons are common environmental pollutants, of which many are mutagenic and carcinogenic. 1-Nitropyrene is the most abundant nitrated polycyclic aromatic hydrocarbon, which causes DNA damage and is carcinogenic in experimental animals. Error-prone translesion synthesis of 1-nitropyrene-derived DNA lesions generates mutations that likely play a role in the etiology of cancer. Here, we report two crystal structures of the human Y-family DNA polymerase iota complexed with the major 1-nitropyrene DNA lesion at the insertion stage, incorporating either dCTP or dATP nucleotide opposite the lesion. Polι maintains the adduct in its active site in two distinct conformations. dCTP forms a Watson-Crick base pair with the adducted guanine and excludes the pyrene ring from the helical DNA, which inhibits replication beyond the lesion. By contrast, the mismatched dATP stacks above the pyrene ring that is intercalated in the helix and achieves a productive conformation for misincorporation. The intra-helical bulky pyrene mimics a base pair in the active site and facilitates adenine misincorporation. By structure-based mutagenesis, we show that the restrictive active site of human polη prevents the intra-helical conformation and A-base misinsertions. This work provides one of the molecular mechanisms for G to T transversions, a signature mutation in human lung cancer.
- Department of Biochemistry, Medical Sciences Building 334, University of Western Ontario, London, ON N6A 5C1, Canada.
Organizational Affiliation: