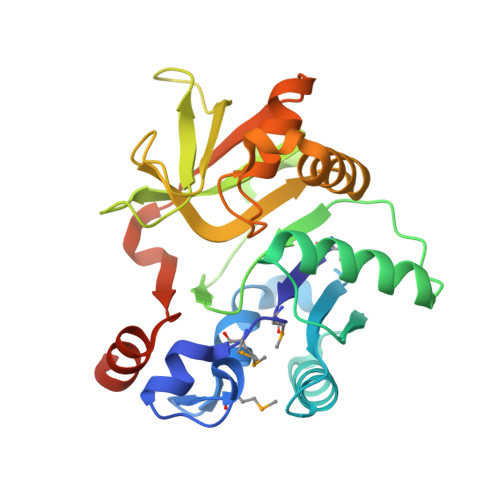
Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9.
Vorobiev, S., Neely, H., Su, M., Seetharaman, J., Mao, M., Xiao, R., Kohan, E., Everett, J.K., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.