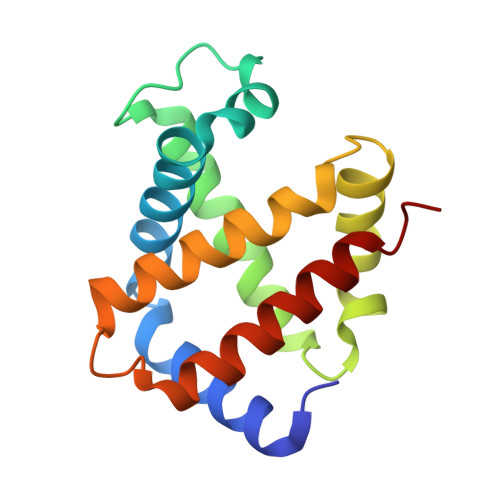
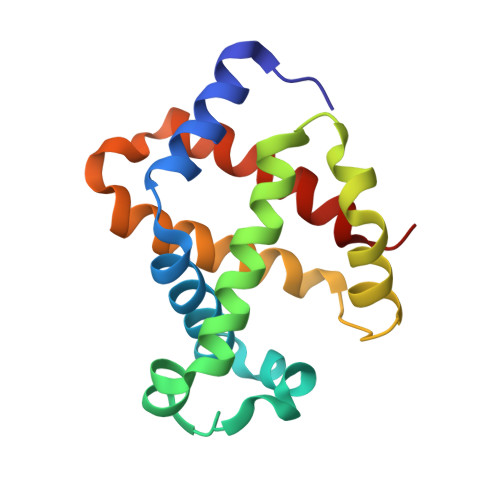
ATP regulation of the ligand-binding properties in temperate and cold-adapted haemoglobins. X-ray structure and ligand-binding kinetics in the sub-Antarctic fish Eleginops maclovinus.
Coppola, D., Abbruzzetti, S., Nicoletti, F., Merlino, A., Gambacurta, A., Giordano, D., Howes, B.D., De Sanctis, G., Vitagliano, L., Bruno, S., di Prisco, G., Mazzarella, L., Smulevich, G., Coletta, M., Viappiani, C., Vergara, A., Verde, C.(2012) Mol Biosyst 8: 3295-3304
- PubMed: 23086282
- DOI: https://doi.org/10.1039/c2mb25210d
- Primary Citation of Related Structures:
4ESA - PubMed Abstract:
The major haemoglobin of the sub-Antarctic fish Eleginops maclovinus was structurally and functionally characterised with the aim to compare molecular environmental adaptations in the O(2)-transport system of sub-Antarctic fishes of the suborder Notothenioidei with those of their high-latitude relatives. Ligand-binding kinetics of the major haemoglobin of E. maclovinus indicated strong stabilisation of the liganded quaternary T state, enhanced in the presence of the physiological allosteric effector ATP, compared to that of high-Antarctic Trematomus bernacchii. The activation enthalpy for O(2) dissociation was dramatically lower than that in T. bernacchii haemoglobin, suggesting remarkable differences in temperature sensitivity and structural changes associated with O(2) release and exit from the protein. The haemoglobin functional properties, together with the X-ray structure of the CO form at 1.49 Å resolution, the first of a temperate notothenioid, strongly support the hypothesis that in E. maclovinus, whose life-style varies according to changes in habitat, the mechanisms that regulate O(2) affinity and the ATP-induced Root effect differ from those of high-Antarctic Notothenioids.
- Institute of Protein Biochemistry, CNR, Via Pietro Castellino 111, I-80131 Naples, Italy.
Organizational Affiliation: