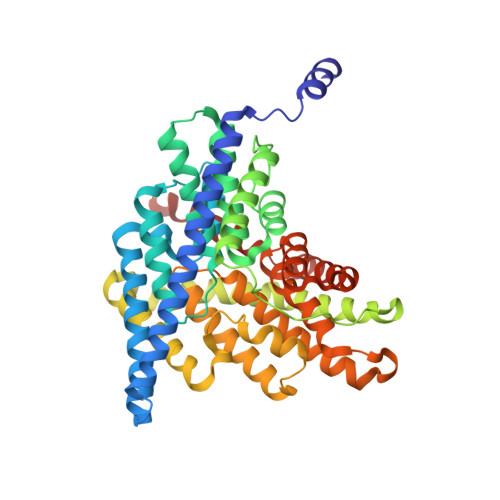
Intracellular proton access in a cl(-)/h(+) antiporter.
Lim, H.H., Shane, T., Miller, C.(2012) PLoS Biol 10: e1001441-e1001441
- PubMed: 23239938
- DOI: https://doi.org/10.1371/journal.pbio.1001441
- Primary Citation of Related Structures:
4ENE - PubMed Abstract:
Chloride-transporting membrane proteins of the CLC family appear in two distinct mechanistic flavors: H(+)-gated Cl(-) channels and Cl(-)/H(+) antiporters. Transmembrane H(+) movement is an essential feature of both types of CLC. X-ray crystal structures of CLC antiporters show the Cl(-) ion pathway through these proteins, but the H(+) pathway is known only inferentially by two conserved glutamate residues that act as way-stations for H(+) in its path through the protein. The extracellular-facing H(+) transfer glutamate becomes directly exposed to aqueous solution during the transport cycle, but the intracellular glutamate E203, Glu(in), is buried within the protein. Two regions, denoted "polar" and "interfacial," at the intracellular surface of the bacterial antiporter CLC-ec1 are examined here as possible pathways by which intracellular aqueous protons gain access to Glu(in). Mutations at multiple residues of the polar region have little effect on antiport rates. In contrast, mutation of E202, a conserved glutamate at the protein-water boundary of the interfacial region, leads to severe slowing of the Cl(-)/H(+) antiport rate. An X-ray crystal structure of E202Y, the most strongly inhibited of these substitutions, shows an aqueous portal leading to Glu(in) physically blocked by cross-subunit interactions; moreover, this mutation has only minimal effect on a monomeric CLC variant, which necessarily lacks such interactions. The several lines of experiments presented argue that E202 acts as a water-organizer that creates a proton conduit connecting intracellular solvent with Glu(in).
- Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts, USA.
Organizational Affiliation: