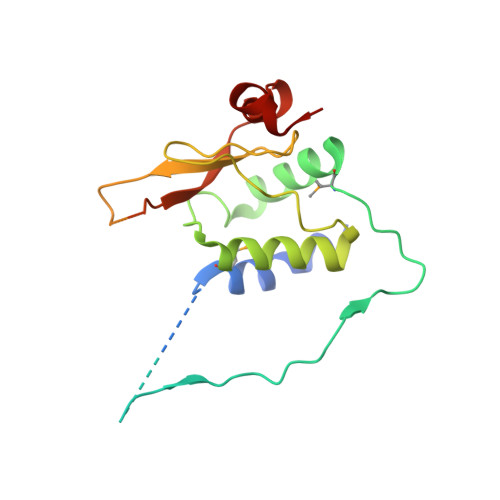
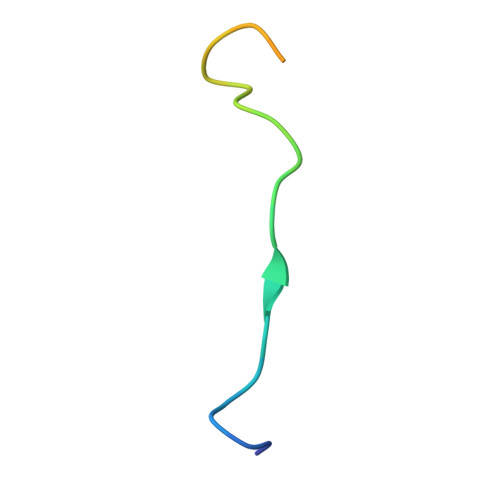
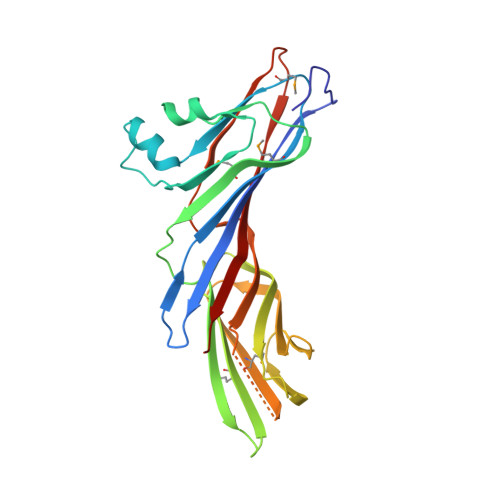
Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef.
Jia, X., Singh, R., Homann, S., Yang, H., Guatelli, J., Xiong, Y.(2012) Nat Struct Mol Biol 19: 701-706
- PubMed: 22705789
- DOI: https://doi.org/10.1038/nsmb.2328
- Primary Citation of Related Structures:
4EMZ, 4EN2 - PubMed Abstract:
The HIV-1 protein Nef inhibits antigen presentation by class I major histocompatibility complex (MHC-I). We determined the mechanism of this activity by solving the crystal structure of a protein complex comprising Nef, the MHC-I cytoplasmic domain (MHC-I CD) and the μ1 subunit of the clathrin adaptor protein complex 1. A ternary, cooperative interaction clamps the MHC-I CD into a narrow binding groove at the Nef-μ1 interface, which encompasses the cargo-recognition site of μ1 and the proline-rich strand of Nef. The Nef C terminus induces a previously unobserved conformational change in μ1, whereas the N terminus binds the Nef core to position it optimally for complex formation. Positively charged patches on μ1 recognize acidic clusters in Nef and MHC-I. The structure shows how Nef functions as a clathrin-associated sorting protein to alter the specificity of host membrane trafficking and enable viral evasion of adaptive immunity.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut, USA.
Organizational Affiliation: