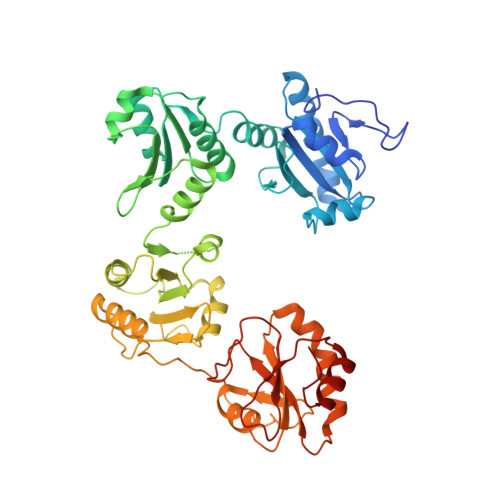
Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Wang, C., Li, W., Ren, J., Fang, J., Ke, H., Gong, W., Feng, W., Wang, C.-C.(2013) Antioxid Redox Signal 19: 44-53
- PubMed: 22657537
- DOI: https://doi.org/10.1089/ars.2012.4630
- Primary Citation of Related Structures:
4EKZ, 4EL1 - PubMed Abstract:
Human protein disulfide isomerase (hPDI) is a key enzyme and a redox-regulated chaperone responsible for oxidative protein folding in the endoplasmic reticulum. This work aims to reveal the molecular mechanism underlying the redox-regulated functions of hPDI by determining the crystal structures of hPDI in different redox states. The structures of hPDI (abb'xa') in both the reduced and oxidized states showed that the four thioredoxin domains of a, b, b', and a' are arranged as a horseshoe shape with two CGHC active sites, respectively, in domains a and a' facing each other at the two ends. In reduced hPDI, domains a, b, and b' line up in the same plane, whereas domain a' twists ∼45° out. The two active sites are 27.6 Å apart. In oxidized hPDI, the four domains are differently organized to stay in the same plane, and the distance between the active sites increases to 40.3 Å. In contrast to the closed conformation of reduced hPDI, oxidized hPDI exists in an open state with more exposed hydrophobic areas and a larger cleft with potential for substrate binding. This is the first report of the high-resolution structures of hPDI containing all four domains in both the reduced and the oxidized states. It reveals the redox-regulated structural dynamic properties of the protein. The redox-regulated open/closed conformational switch of hPDI endows the protein with versatile target-binding capacities for its enzymatic and chaperone functions.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: