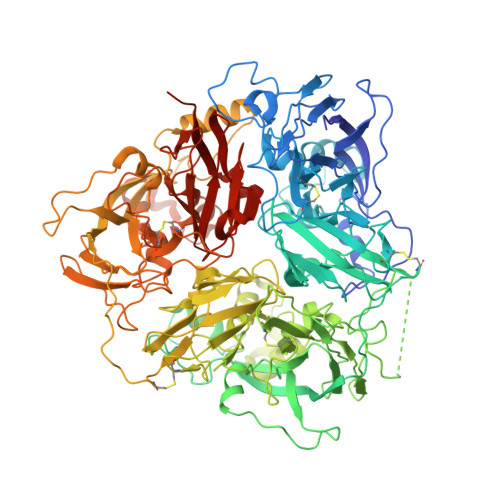
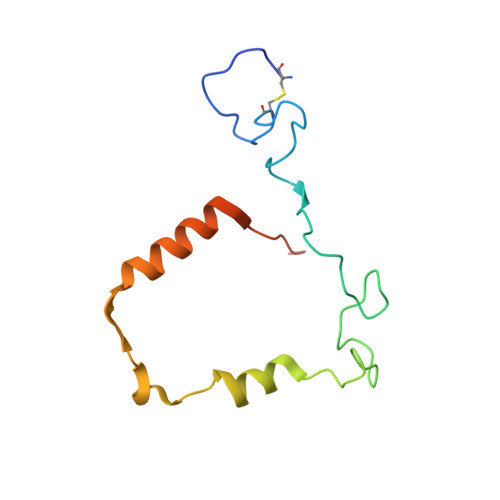
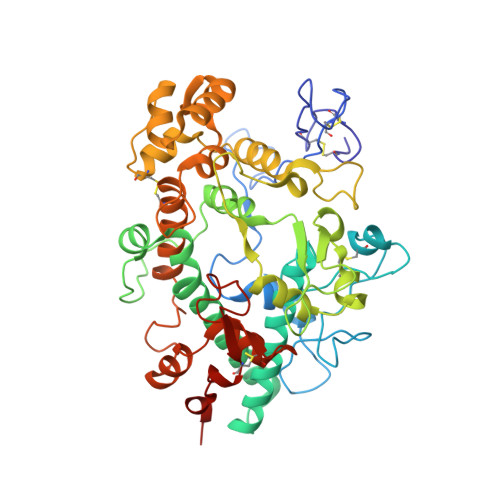
Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Samygina, V.R., Sokolov, A.V., Bourenkov, G., Petoukhov, M.V., Pulina, M.O., Zakharova, E.T., Vasilyev, V.B., Bartunik, H., Svergun, D.I.(2013) PLoS One 8: e67145-e67145
- PubMed: 23843990
- DOI: https://doi.org/10.1371/journal.pone.0067145
- Primary Citation of Related Structures:
4EJX, 4ENZ - PubMed Abstract:
Copper-containing ferroxidase ceruloplasmin (Cp) forms binary and ternary complexes with cationic proteins lactoferrin (Lf) and myeloperoxidase (Mpo) during inflammation. We present an X-ray crystal structure of a 2Cp-Mpo complex at 4.7 Å resolution. This structure allows one to identify major protein-protein interaction areas and provides an explanation for a competitive inhibition of Mpo by Cp and for the activation of p-phenylenediamine oxidation by Mpo. Small angle X-ray scattering was employed to construct low-resolution models of the Cp-Lf complex and, for the first time, of the ternary 2Cp-2Lf-Mpo complex in solution. The SAXS-based model of Cp-Lf supports the predicted 1:1 stoichiometry of the complex and demonstrates that both lobes of Lf contact domains 1 and 6 of Cp. The 2Cp-2Lf-Mpo SAXS model reveals the absence of interaction between Mpo and Lf in the ternary complex, so Cp can serve as a mediator of protein interactions in complex architecture. Mpo protects antioxidant properties of Cp by isolating its sensitive loop from proteases. The latter is important for incorporation of Fe(3+) into Lf, which activates ferroxidase activity of Cp and precludes oxidation of Cp substrates. Our models provide the structural basis for possible regulatory role of these complexes in preventing iron-induced oxidative damage.
- Institute of Crystallography RAS, Moscow, Russia. vsamygina@cicbiogune.es
Organizational Affiliation: