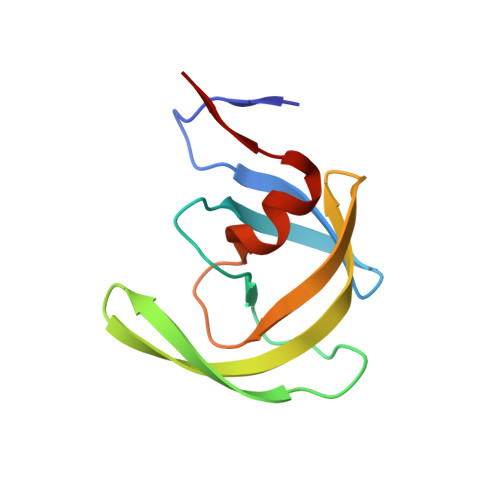
Small molecule regulation of protein conformation by binding in the Flap of HIV protease.
Tiefenbrunn, T., Forli, S., Baksh, M.M., Chang, M.W., Happer, M., Lin, Y.C., Perryman, A.L., Rhee, J.K., Torbett, B.E., Olson, A.J., Elder, J.H., Finn, M.G., Stout, C.D.(2013) ACS Chem Biol 8: 1223-1231
- PubMed: 23540839
- DOI: https://doi.org/10.1021/cb300611p
- Primary Citation of Related Structures:
4EJ8, 4EJD, 4EJK, 4EJL - PubMed Abstract:
The fragment indole-6-carboxylic acid (1F1), previously identified as a flap site binder in a fragment-based screen against HIV protease (PR), has been cocrystallized with pepstatin-inhibited PR and with apo-PR. Another fragment, 3-indolepropionic acid (1F1-N), predicted by AutoDock calculations and confirmed in a novel inhibition of nucleation crystallization assay, exploits the same interactions in the flap site in two crystal structures. Both 1F1 and 1F1-N bind to the closed form of apo-PR and to pepstatin:PR. In solution, 1F1 and 1F1-N raise the Tm of apo-PR by 3.5-5 °C as assayed by differential scanning fluorimetry (DSF) and show equivalent low-micromolar binding constants to both apo-PR and pepstatin:PR, assayed by backscattering interferometry (BSI). The observed signal intensities in BSI are greater for each fragment upon binding to apo-PR than to pepstatin-bound PR, consistent with greater conformational change in the former binding event. Together, these data indicate that fragment binding in the flap site favors a closed conformation of HIV PR.
- Deparatment of Integrative Structural and Computational Biology, ‡Department of Chemistry, §Department of Molecular and Experimental Medicine, ∥Department of Immunology and Microbial Science, The Scripps Research Institute , 10550 N. Torrey Pines Rd., La Jolla, California 92037, United States.
Organizational Affiliation: