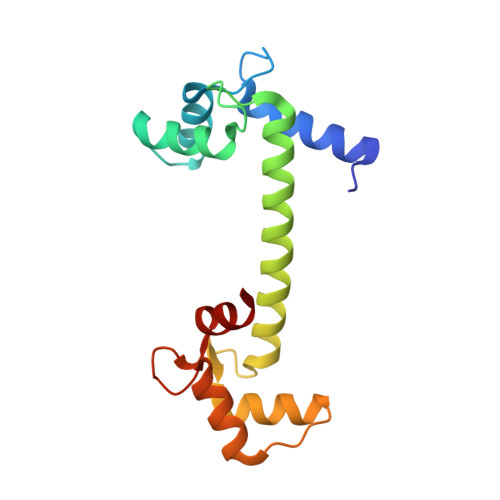
Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
Liu, Y., Zheng, X., Mueller, G.A., Sobhany, M., Derose, E.F., Zhang, Y., London, R.E., Birnbaumer, L.(2012) J Biological Chem 287: 43030-43041
- PubMed: 23109337
- DOI: https://doi.org/10.1074/jbc.M112.380964
- Primary Citation of Related Structures:
4EHQ - PubMed Abstract:
Orai1 is a plasma membrane protein that in its tetrameric form is responsible for calcium influx from the extracellular environment into the cytosol in response to interaction with the Ca(2+)-depletion sensor STIM1. This is followed by a fast Ca(2+)·calmodulin (CaM)-dependent inhibition, resulting from CaM binding to an Orai1 region called the calmodulin binding domain (CMBD). The interaction between Orai1 and CaM at the atomic level remains unknown. Here, we report the crystal structure of a CaM·Orai1-CMBD complex showing one CMBD bound to the C-terminal lobe of CaM, differing from other CaM-target protein complexes, in which both N- and C-terminal lobes of CaM (CaM-N and CaM-C) are involved in target binding. Orai1-CMBD binds CaM-C mainly through hydrophobic interactions, primarily involving residue Trp(76) of Orai1-CMBD, which interacts with the hydrophobic pocket of CaM-C. However, NMR data, isothermal titration calorimetry data, and pulldown assays indicated that CaM-N and CaM-C both can bind Orai1-CMBD, with CaM-N having ∼4 times weaker affinity than CaM-C. Pulldown assays of a Orai1-CMBD(W76E) mutant, gel filtration chromatography data, and NOE signals indicated that CaM-N and CaM-C can each bind one Orai1-CMBD. Thus our studies support an unusual, extended 1:2 binding mode of CaM to Orai1-CMBDs, and quantify the affinity of Orai1 for CaM. We propose a two-step mechanism for CaM-dependent Orai1 inactivation initiated by binding of the C-lobe of CaM to the CMBD of one Orai1 followed by the binding of the N-lobe of CaM to the CMBD of a neighboring Orai1.
- Laboratory of Neurobiology, NIEHS, National Institutes of Health, Research Triangle Park, North Carolina 27709, USA. liuy3@niehs.nih.gov
Organizational Affiliation: