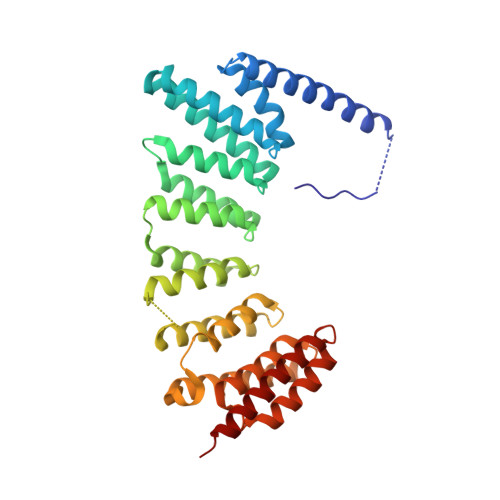
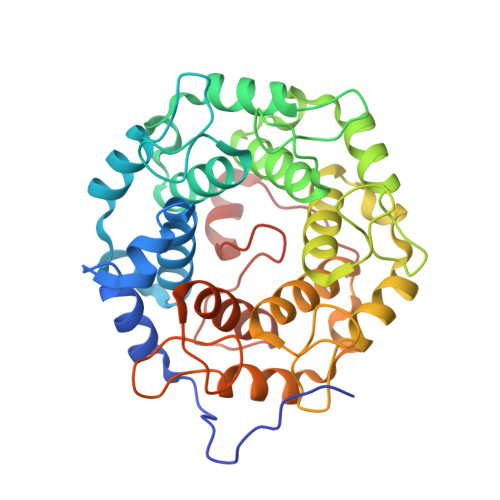
Psoromic Acid is a Selective and Covalent Rab-Prenylation Inhibitor Targeting Autoinhibited RabGGTase
Deraeve, C., Guo, Z., Bon, R.S., Blankenfeldt, W., Dilucrezia, R., Wolf, A., Menninger, S., Stigter, E.A., Wetzel, S., Choidas, A., Alexandrov, K., Waldmann, H., Goody, R.S., Wu, Y.W.(2012) J Am Chem Soc 134: 7384-7391
- PubMed: 22480322
- DOI: https://doi.org/10.1021/ja211305j
- Primary Citation of Related Structures:
4EHM - PubMed Abstract:
Post-translational attachment of geranylgeranyl isoprenoids to Rab GTPases, the key organizers of intracellular vesicular transport, is essential for their function. Rab geranylgeranyl transferase (RabGGTase) is responsible for prenylation of Rab proteins. Recently, RabGGTase inhibitors have been proposed to be potential therapeutics for treatment of cancer and osteoporosis. However, the development of RabGGTase selective inhibitors is complicated by its structural and functional similarity to other protein prenyltransferases. Herein we report identification of the natural product psoromic acid (PA) that potently and selectively inhibits RabGGTase with an IC(50) of 1.3 μM. Structure-activity relationship analysis suggested a minimal structure involving the depsidone core with a 3-hydroxyl and 4-aldehyde motif for binding to RabGGTase. Analysis of the crystal structure of the RabGGTase:PA complex revealed that PA forms largely hydrophobic interactions with the isoprenoid binding site of RabGGTase and that it attaches covalently to the N-terminus of the α subunit. We found that in contrast to other protein prenyltransferases, RabGGTase is autoinhibited through N-terminal (α)His2 coordination with the catalytic zinc ion. Mutation of (α)His dramatically enhances the reaction rate, indicating that the activity of RabGGTase is likely regulated in vivo. The covalent binding of PA to the N-terminus of the RabGGTase α subunit seems to potentiate its interaction with the active site and explains the selectivity of PA for RabGGTase. Therefore, psoromic acid provides a new starting point for the development of selective RabGGTase inhibitors.
- Department of Chemical Biology, Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: