Crystal structure of DNA ligase
Wang, T., Charifson, P., Xu, W., Wei, Y.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA ligase | 331 | Enterococcus faecalis | Mutation(s): 0 EC: 6.5.1.2 | 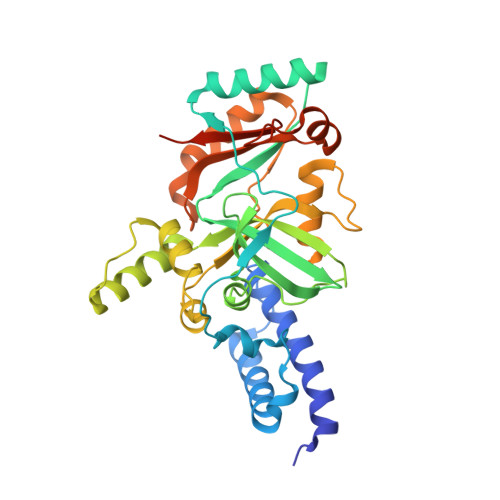 | |
UniProt | |||||
Find proteins for Q837V6 (Enterococcus faecalis (strain ATCC 700802 / V583)) Explore Q837V6 Go to UniProtKB: Q837V6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q837V6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NMN Query on NMN | D [auth A] | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE C11 H16 N2 O8 P DAYLJWODMCOQEW-TURQNECASA-O |  | ||
| 0OW Query on 0OW | E [auth A] | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide C15 H22 N4 O4 GOYBCUMANOWCAY-VHSXEESVSA-N |  | ||
| SO4 Query on SO4 | B [auth A], C [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 90.13 | α = 90 |
| b = 86.01 | β = 100.87 |
| c = 56.84 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| CNX | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |