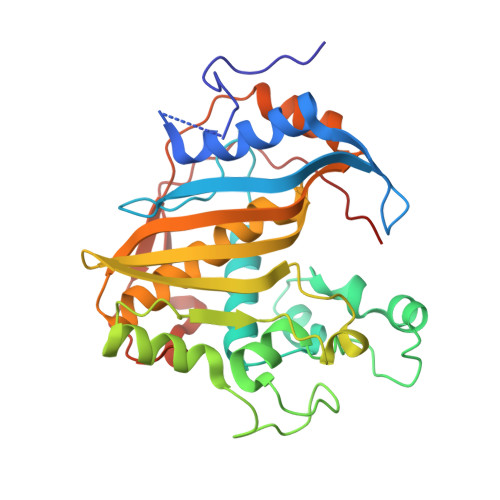
Crystal structure of mouse thymidylate synthase in tertiary complex with dUMP and raltitrexed reveals N-terminus architecture and two different active site conformations.
Dowiercial, A., Wilk, P., Rypniewski, W., Rode, W., Jarmula, A.(2014) Biomed Res Int 2014: 945803
- PubMed: 24995339
- DOI: https://doi.org/10.1155/2014/945803
- Primary Citation of Related Structures:
4EB4 - PubMed Abstract:
The crystal structure of mouse thymidylate synthase (mTS) in complex with substrate dUMP and antifolate inhibitor Raltitrexed is reported. The structure reveals, for the first time in the group of mammalian TS structures, a well-ordered segment of 13 N-terminal amino acids, whose ordered conformation is stabilized due to specific crystal packing. The structure consists of two homodimers, differing in conformation, one being more closed (dimer AB) and thus supporting tighter binding of ligands, and the other being more open (dimer CD) and thus allowing weaker binding of ligands. This difference indicates an asymmetrical effect of the binding of Raltitrexed to two independent mTS molecules. Conformational changes leading to a ligand-induced closing of the active site cleft are observed by comparing the crystal structures of mTS in three different states along the catalytic pathway: ligand-free, dUMP-bound, and dUMP- and Raltitrexed-bound. Possible interaction routes between hydrophobic residues of the mTS protein N-terminal segment and the active site are also discussed.
- Nencki Institute of Experimental Biology, Polish Academy of Sciences, 3 Pasteur Street, 02-093 Warsaw, Poland.
Organizational Affiliation: