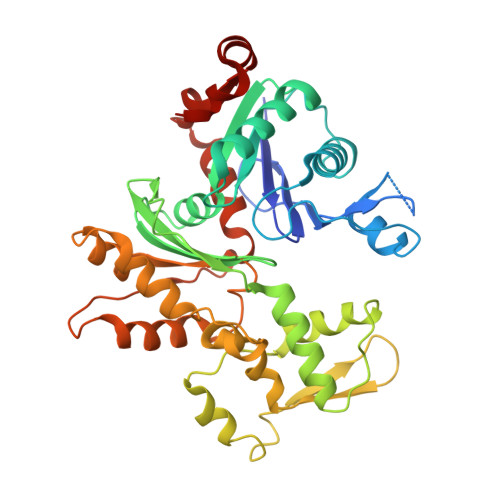
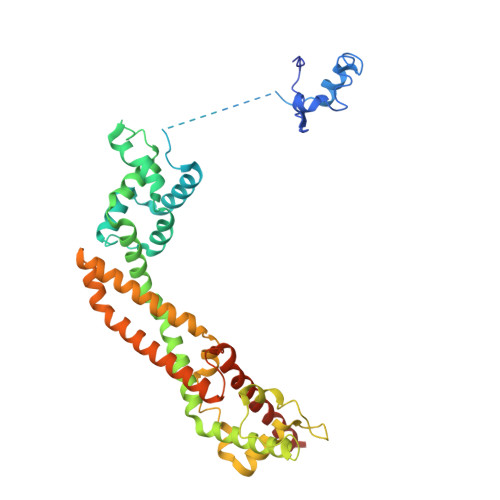
FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation.
Thompson, M.E., Heimsath, E.G., Gauvin, T.J., Higgs, H.N., Kull, F.J.(2013) Nat Struct Mol Biol 20: 111-118
- PubMed: 23222643
- DOI: https://doi.org/10.1038/nsmb.2462
- Primary Citation of Related Structures:
4EAH - PubMed Abstract:
Formins are actin-assembly factors that act in a variety of actin-based processes. The conserved formin homology 2 (FH2) domain promotes filament nucleation and influences elongation through interaction with the barbed end. FMNL3 is a formin that induces assembly of filopodia but whose FH2 domain is a poor nucleator. The 3.4-Å structure of a mouse FMNL3 FH2 dimer in complex with tetramethylrhodamine-actin uncovers details of formin-regulated actin elongation. We observe distinct FH2 actin-binding regions; interactions in the knob and coiled-coil subdomains are necessary for actin binding, whereas those in the lasso-post interface are important for the stepping mechanism. Biochemical and cellular experiments test the importance of individual residues for function. This structure provides details for FH2-mediated filament elongation by processive capping and supports a model in which C-terminal non-FH2 residues of FMNL3 are required to stabilize the filament nucleus.
- Department of Biochemistry, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire, USA.
Organizational Affiliation: