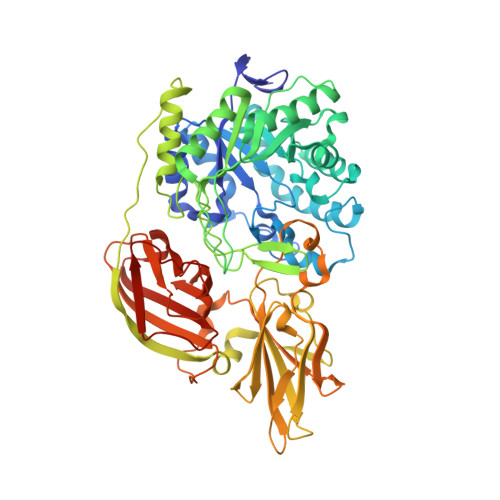
Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
Cheng, W., Wang, L., Jiang, Y.L., Bai, X.H., Chu, J., Li, Q., Yu, G., Liang, Q.L., Zhou, C.Z., Chen, Y.X.(2012) J Biological Chem 287: 22910-22918
- PubMed: 22593580
- DOI: https://doi.org/10.1074/jbc.M112.367128
- Primary Citation of Related Structures:
4E8C, 4E8D - PubMed Abstract:
The surface-exposed β-galactosidase BgaC from Streptococcus pneumoniae was reported to be a virulence factor because of its specific hydrolysis activity toward the β(1,3)-linked galactose and N-acetylglucosamine (Galβ(1,3)NAG) moiety of oligosaccharides on the host molecules. Here we report the crystal structure of BgaC at 1.8 Å and its complex with galactose at 1.95 Å. At pH 5.5-8.0, BgaC exists as a stable homodimer, each subunit of which consists of three distinct domains: a catalytic domain of a classic (β/α)(8) TIM barrel, followed by two all-β domains (ABDs) of unknown function. The side walls of the TIM β-barrel and a loop extended from the first ABD constitute the active site. Superposition of the galactose-complexed structure to the apo-form revealed significant conformational changes of residues Trp-243 and Tyr-455. Simulation of a putative substrate entrance tunnel and modeling of a complex structure with Galβ(1,3)NAG enabled us to assign three key residues to the specific catalysis. Site-directed mutagenesis in combination with activity assays further proved that residues Trp-240 and Tyr-455 contribute to stabilizing the N-acetylglucosamine moiety, whereas Trp-243 is critical for fixing the galactose ring. Moreover, we propose that BgaC and other galactosidases in the GH-35 family share a common domain organization and a conserved substrate-determinant aromatic residue protruding from the second domain.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: