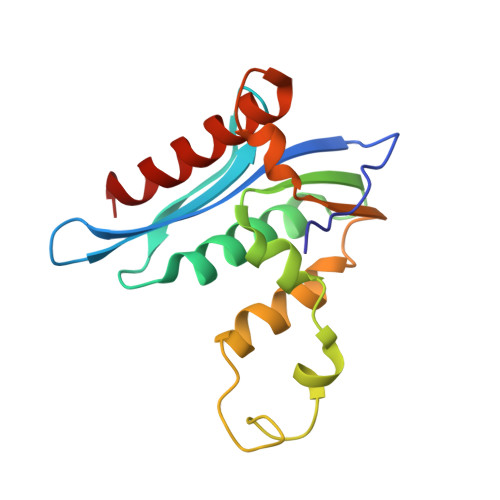
Crystal structure of xenotropic murine leukaemia virus-related virus (XMRV) ribonuclease H
Kim, J.H., Kang, S., Jung, S.K., Yu, K.R., Chung, S.J., Chung, B.H., Erikson, R.L., Kim, B.Y., Kim, S.J.(2012) Biosci Rep 32: 455-463
- PubMed: 22724525
- DOI: https://doi.org/10.1042/BSR20120028
- Primary Citation of Related Structures:
4E89 - PubMed Abstract:
RNase H (retroviral ribonuclease H) cleaves the phosphate backbone of the RNA template within an RNA/DNA hybrid to complete the synthesis of double-stranded viral DNA. In the present study we have determined the complete structure of the RNase H domain from XMRV (xenotropic murine leukaemia virus-related virus) RT (reverse transcriptase). The basic protrusion motif of the XMRV RNase H domain is folded as a short helix and an adjacent highly bent loop. Structural superposition and subsequent mutagenesis experiments suggest that the basic protrusion motif plays a role in direct binding to the major groove in RNA/DNA hybrid, as well as in establishing the co-ordination among modules in RT necessary for proper function.
- Medical Proteomics Research Center, Korea Research Institute of Bioscience and Biotechnology, Yuseong-Gu, Daejeon, Republic of Korea.
Organizational Affiliation: