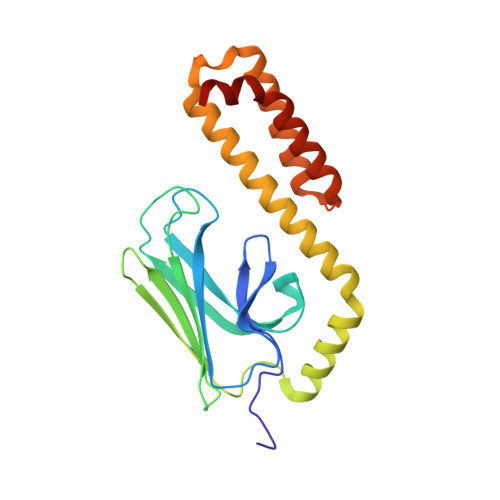
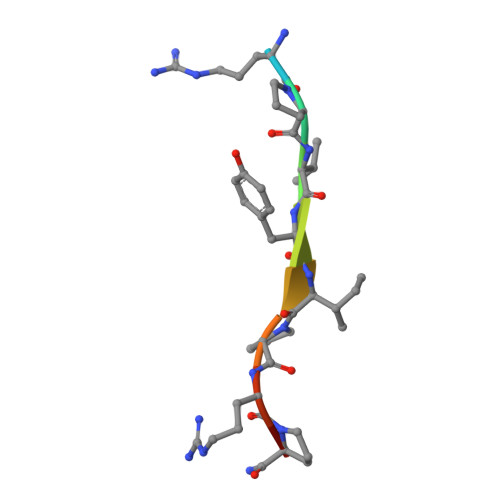
Api88 is a novel antibacterial designer Peptide to treat systemic infections with multidrug-resistant gram-negative pathogens.
Czihal, P., Knappe, D., Fritsche, S., Zahn, M., Berthold, N., Piantavigna, S., Muller, U., Van Dorpe, S., Herth, N., Binas, A., Kohler, G., De Spiegeleer, B., Martin, L.L., Nolte, O., Strater, N., Alber, G., Hoffmann, R.(2012) ACS Chem Biol 7: 1281-1291
- PubMed: 22594381
- DOI: https://doi.org/10.1021/cb300063v
- Primary Citation of Related Structures:
4E81 - PubMed Abstract:
The emergence of multiple-drug-resistant (MDR) bacterial pathogens in hospitals (nosocomial infections) presents a global threat of growing importance, especially for Gram-negative bacteria with extended spectrum β-lactamase (ESBL) or the novel New Delhi metallo-β-lactamase 1 (NDM-1) resistance. Starting from the antibacterial peptide apidaecin 1b, we have optimized the sequence to treat systemic infections with the most threatening human pathogens, such as Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. The lead compound Api88 enters bacteria without lytic effects at the membrane and inhibits chaperone DnaK at the substrate binding domain with a K(D) of 5 μmol/L. The Api88-DnaK crystal structure revealed that Api88 binds with a seven residue long sequence (PVYIPRP), in two different modes. Mice did not show any sign of toxicity when Api88 was injected four times intraperitoneally at a dose of 40 mg/kg body weight (BW) within 24 h, whereas three injections of 1.25 mg/kg BW and 5 mg/kg BW were sufficient to rescue all animals in lethal sepsis models using pathogenic E. coli strains ATCC 25922 and Neumann, respectively. Radioactive labeling showed that Api88 enters all organs investigated including the brain and is cleared through both the liver and kidneys at similar rates. In conclusion, Api88 is a novel, highly promising, 18-residue peptide lead compound with favorable in vitro and in vivo properties including a promising safety margin.
- Institute of Bioanalytical Chemistry, Faculty of Chemistry and Mineralogy, College of Veterinary Medicine, Universität Leipzig, Germany.
Organizational Affiliation: