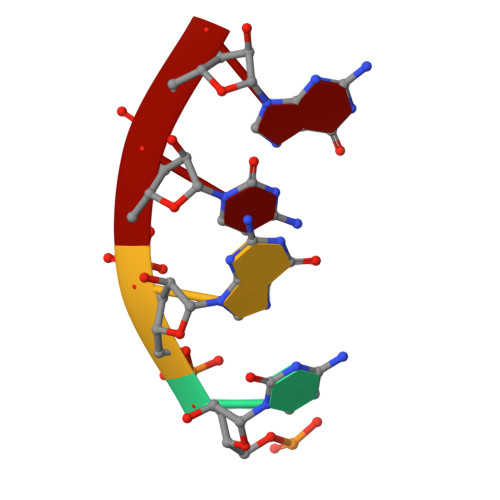
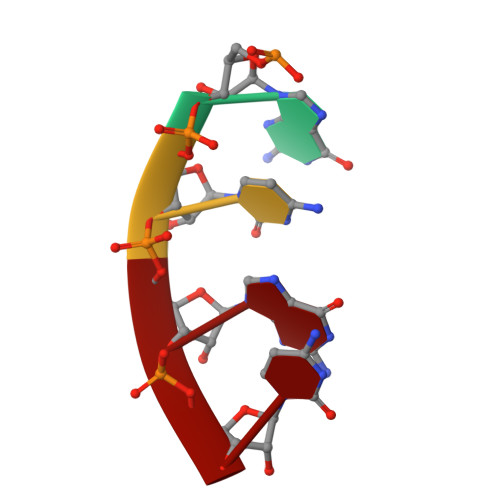
Structural dynamics of double-helical RNAs composed of CUG/CUG- and CUG/CGG-repeats.
Tamjar, J., Katorcha, E., Popov, A., Malinina, L.(2012) J Biomol Struct Dyn 30: 505-523
- PubMed: 22731704
- DOI: https://doi.org/10.1080/07391102.2012.687517
- Primary Citation of Related Structures:
4E48, 4E5C, 4E6B - PubMed Abstract:
Human genetic trinucleotide repeat expansion diseases (TREDs) are characterized by triplet repeat expansions, most frequently found as CNG-tracts in genome. At RNA level, such expansions suggestively result in formation of double-helical hairpins that become a potential source for small RNAs involved in RNA interference (RNAi). Here, we present three crystal structures of RNA fragments composed of triplet repeats CUG and CGG/CUG, as well as two crystal structures of same triplets in a protein-bound state. We show that both 20mer pG(CUG)(6)C and 19mer pGG(CGG)(3)(CUG)(2)CC form A-RNA duplexes, in which U·U or G·U mismatches are flanked/stabilized by two consecutive Watson-Crick G·C base pairs resulting in high-stacking GpC steps in every third position of the duplex. Despite interruption of this regularity in another 19mer, p(CGG)(3)C(CUG)(3), the oligonucleotide still forms regular double-helical structure, characterized, however, by 12 bp (rather than 11 bp) per turn. Analysis of newly determined molecular structures reveals the dynamic aspects of U·U and G·U mismatching within CNG-repetitive A-RNA and in a protein-bound state, as well as identifies an additional mode of U·U pairing essential for its dynamics and sheds the light on possible role of regularity of trinucleotide repeats for double-helical RNA structure. Findings are important for understanding the structural behavior of CNG-repetitive RNA double helices implicated in TREDs.
- Structural Biology Unit, CIC bioGUNE, Technology Park of Bizkaia, Derio-Bilbao 48160, Spain.
Organizational Affiliation: