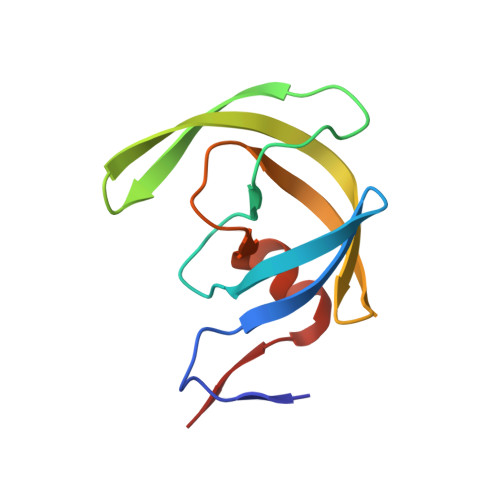
Fragment-based screen against HIV protease.
Perryman, A.L., Zhang, Q., Soutter, H.H., Rosenfeld, R., McRee, D.E., Olson, A.J., Elder, J.E., Stout, C.D.(2010) Chem Biol Drug Des 75: 257-268
- PubMed: 20659109
- DOI: https://doi.org/10.1111/j.1747-0285.2009.00943.x
- Primary Citation of Related Structures:
3KF0, 3KFN, 3KFP, 3KFR, 3KFS, 4E43 - PubMed Abstract:
We have employed a fragment-based screen against wild-type (NL4-3) HIV protease (PR) using the Active Sight fragment library and X-ray crystallography. The experiments reveal two new binding sites for small molecules. PR was co-crystallized with fragments, or crystals were soaked in fragment solutions, using five crystal forms, and 378 data sets were collected to 2.3-1.3 A resolution. Fragment binding induces a distinct conformation and specific crystal form of TL-3 inhibited PR during co-crystallization. One fragment, 2-methylcyclohexanol, binds in the 'exo site' adjacent to the Gly(16)Gly(17)Gln(18)loop where the amide of Gly(17)is a specific hydrogen bond donor, and hydrophobic contacts occur with the side chains of Lys(14)and Leu(63). Another fragment, indole-6-carboxylic acid, binds on the 'outside/top of the flap' via hydrophobic contacts with Trp(42), Pro(44), Met(46), and Lys(55), a hydrogen bond with Val(56), and a salt-bridge with Arg(57). 2-acetyl-benzothiophene also binds at this site. This study is the first fragment-based crystallographic screen against HIV PR, and the first time that fragments were screened against an inhibitor-bound drug target to search for compounds that both bind to novel sites and stabilize the inhibited conformation of the target.
- Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Rd., La Jolla, CA 92037, USA.
Organizational Affiliation: