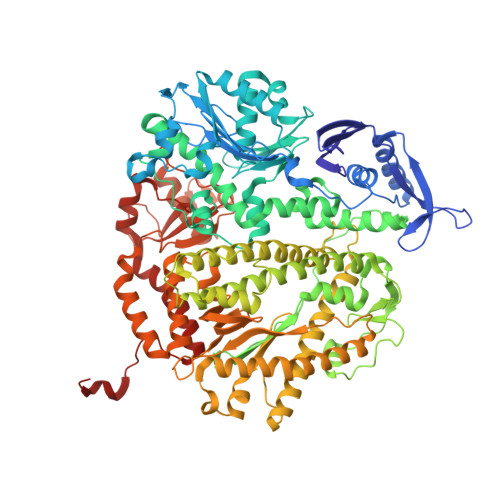
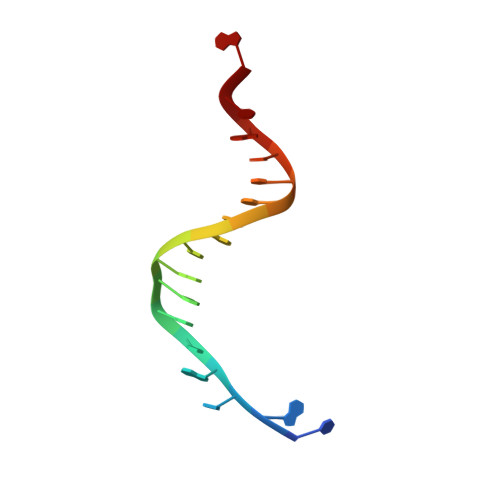
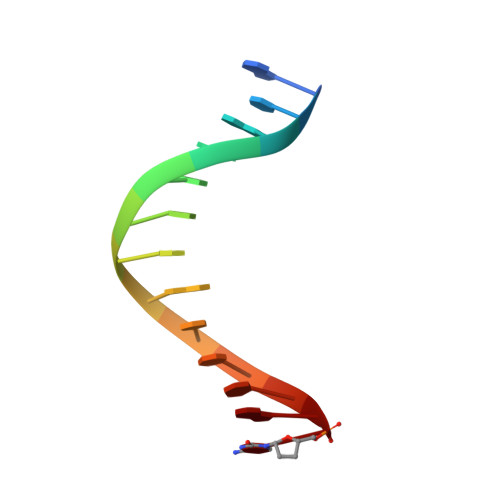
Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Xia, S., Christian, T.D., Wang, J., Konigsberg, W.H.(2012) Biochemistry 51: 4343-4353
- PubMed: 22571765
- DOI: https://doi.org/10.1021/bi300416z
- Primary Citation of Related Structures:
4DU1, 4DU3, 4DU4, 4E3S - PubMed Abstract:
Minor groove hydrogen bonding (HB) interactions between DNA polymerases (pols) and N3 of purines or O2 of pyrimidines have been proposed to be essential for DNA synthesis from results obtained using various nucleoside analogues lacking the N3 or O2 contacts that interfered with primer extension. Because there has been no direct structural evidence to support this proposal, we decided to evaluate the contribution of minor groove HB interactions with family B pols. We have used RB69 DNA pol and 3-deaza-2'-deoxyadenosine (3DA), an analogue of 2-deoxyadenosine, which has the same HB pattern opposite T but with N3 replaced with a carbon atom. We then determined pre-steady-state kinetic parameters for the insertion of dAMP opposite dT using primer/templates (P/T)-containing 3DA. We also determined three structures of ternary complexes with 3DA at various positions in the duplex DNA substrate. We found that the incorporation efficiency of dAMP opposite dT decreased 10(2)-10(3)-fold even when only one minor groove HB interaction was missing. Our structures show that the HB pattern and base pair geometry of 3DA/dT is exactly the same as those of dA/dT, which makes 3DA an optimal analogue for probing minor groove HB interactions between a DNA polymerase and a nucleobase. In addition, our structures provide a rationale for the observed 10(2)-10(3)-fold decrease in the rate of nucleotide incorporation. The minor groove HB interactions between position n - 2 of the primer strand and RB69pol fix the rotomer conformations of the K706 and D621 side chains, as well as the position of metal ion A and its coordinating ligands, so that they are in the optinal orientation for DNA synthesis.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA.
Organizational Affiliation: