X-ray crystal structure of human IKK2 in an active conformation
Polley, S., Huang, D.B., Hauenstein, A.V., Fusco, A.J., Zhong, X., Vu, D., Hoffmann, A., Verma, I.V., Ghosh, G., Huaford, T.(2013) PLoS Biol
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2013) PLoS Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Inhibitor of nuclear factor kappa-B kinase subunit beta | 669 | Homo sapiens | Mutation(s): 2 Gene Names: IKBKB, IKKB EC: 2.7.11.10 (PDB Primary Data), 2.7.11.1 (UniProt) | 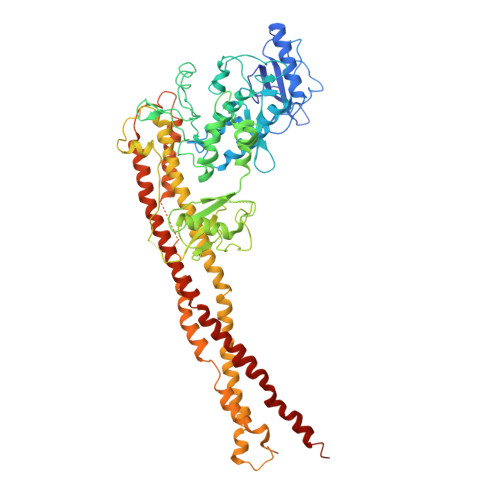 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O14920 (Homo sapiens) Explore O14920 Go to UniProtKB: O14920 | |||||
PHAROS: O14920 GTEx: ENSG00000104365 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O14920 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 170.81 | α = 90 |
| b = 170.81 | β = 90 |
| c = 509.56 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| CNS | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |