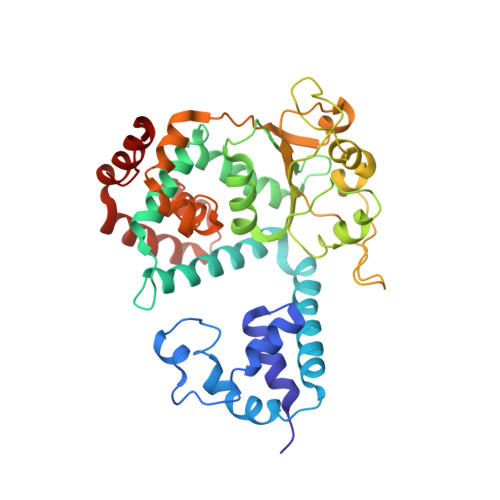
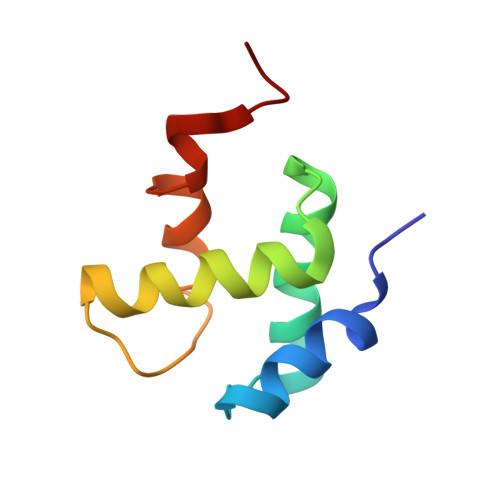
Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
Zhou, B., Arnett, D.R., Yu, X., Brewster, A., Sowd, G.A., Xie, C.L., Vila, S., Gai, D., Fanning, E., Chen, X.S.(2012) J Biological Chem 287: 26854-26866
- PubMed: 22700977
- DOI: https://doi.org/10.1074/jbc.M112.363655
- Primary Citation of Related Structures:
4E2I - PubMed Abstract:
DNA polymerase α-primase (Pol-prim) plays an essential role in eukaryotic DNA replication, initiating synthesis of the leading strand and of each Okazaki fragment on the lagging strand. Pol-prim is composed of a primase heterodimer that synthesizes an RNA primer, a DNA polymerase subunit that extends the primer, and a regulatory B-subunit (p68) without apparent enzymatic activity. Pol-prim is thought to interact with eukaryotic replicative helicases, forming a dynamic multiprotein assembly that displays primosome activity. At least three subunits of Pol-prim interact physically with the hexameric replicative helicase SV40 large T antigen, constituting a simple primosome that is active in vitro. However, structural understanding of these interactions and their role in viral chromatin replication in vivo remains incomplete. Here, we report the detailed large T antigen-p68 interface, as revealed in a co-crystal structure and validated by site-directed mutagenesis, and we demonstrate its functional importance in activating the SV40 primosome in cell-free reactions with purified Pol-prim, as well as in monkey cells in vivo.
- Department of Molecular and Computational Biology, University of Southern California, Los Angeles, California 90089, USA.
Organizational Affiliation: