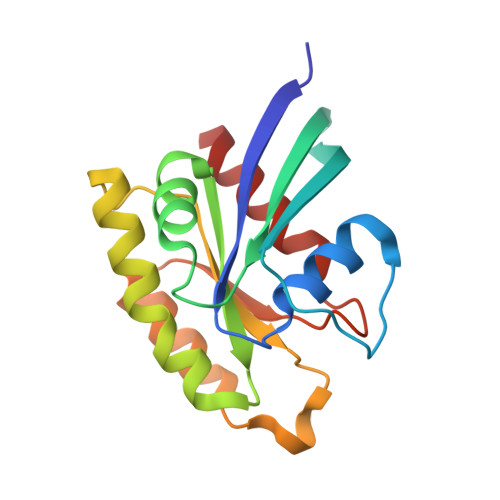
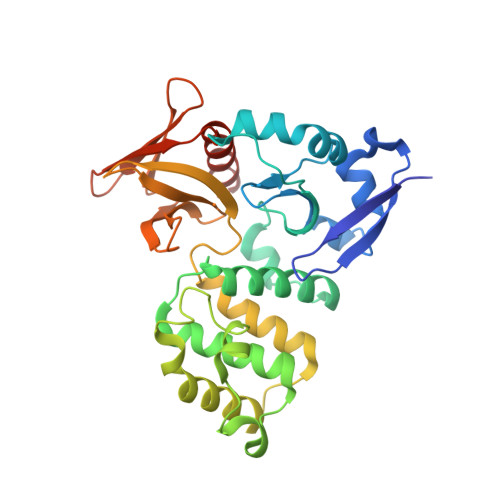
Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
Li, X., Zhang, R., Draheim, K.M., Liu, W., Calderwood, D.A., Boggon, T.J.(2012) J Biological Chem 287: 22317-22327
- PubMed: 22577140
- DOI: https://doi.org/10.1074/jbc.M112.361295
- Primary Citation of Related Structures:
4DXA - PubMed Abstract:
Cerebral cavernous malformations (CCMs) affect 0.1-0.5% of the population resulting in leaky vasculature and severe neurological defects. KRIT1 (Krev interaction trapped-1) mutations associate with ∼40% of familial CCMs. KRIT1 is an effector of Ras-related protein 1 (Rap1) GTPase. Rap1 relocalizes KRIT1 from microtubules to cell membranes to impact integrin activation, potentially important for CCM pathology. We report the 1.95 Å co-crystal structure of KRIT1 FERM domain in complex with Rap1. Rap1-KRIT1 interaction encompasses an extended surface, including Rap1 Switch I and II and KRIT1 FERM F1 and F2 lobes. Rap1 binds KRIT1-F1 lobe using a GTPase-ubiquitin-like fold interaction but binds KRIT1-F2 lobe by a novel interaction. Point mutagenesis confirms the interaction. High similarity between KRIT1-F2/F3 and talin is revealed. Additionally, the mechanism for FERM domains acting as GTPase effectors is suggested. Finally, structure-based alignment of each lobe suggests classification of FERM domains as ERM-like and TMFK-like (talin-myosin-FAK-KRIT-like) and that FERM lobes resemble domain "modules."
- Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut 06520, USA.
Organizational Affiliation: