Invariant gly residue is important for alpha-defensin folding, dimerization, and function: a case study of the human neutrophil alpha-defensin HNP1
Zhao, L., Ericksen, B., Wu, X., Zhan, C., Yuan, W., Li, X., Pazgier, M., Lu, W.(2012) J Biological Chem 287: 18900-18912
- PubMed: 22496447
- DOI: https://doi.org/10.1074/jbc.M112.355255
- Primary Citation of Related Structures:
4DU0 - PubMed Abstract:
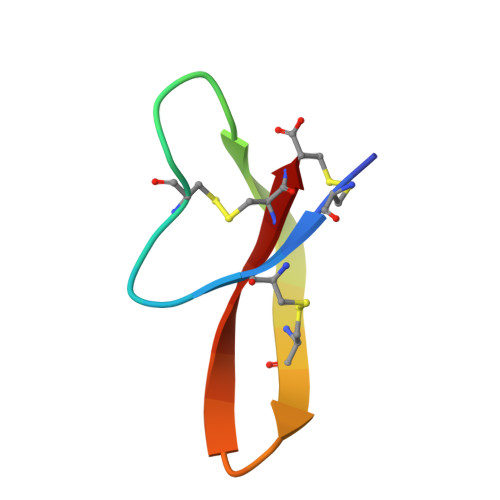
The human α-defensins (HNP) are synthesized in vivo as inactive prodefensins, and contain a conserved glycine, Gly(17), which is part of a β-bulge structure. It had previously been shown that the glycine main chain torsion angles are in a D-configuration, and that d-amino acids but not L-alanine could be substituted at that position to yield correctly folded peptides without the help of a prodomain. In this study, the glycine to L-alanine mutant defensin was synthesized in the form of a prodefensin using native chemical ligation. The ligation product folded correctly and yielded an active peptide upon CNBr cleavage. The L-Ala(17)-HNP1 crystal structure depicted a β-bulge identical to wild-type HNP1. However, dimerization was perturbed, causing one monomer to tilt with respect to the other in a dimerization model. Inhibitory activity against the anthrax lethal factor showed a 2-fold reduction relative to wild-type HNP1 as measured by the inhibitory concentration IC(50). Self-association was slightly reduced, as detected by surface plasmon resonance measurements. According to the results of the virtual colony count assay, the antibacterial activity against Escherichia coli, Staphylococcus aureus, and Bacillus cereus exhibited a less than 2-fold reduction in virtual lethal dose values. Prodefensins with two other L-amino acid substitutions, Arg and Phe, at the same position did not fold, indicating that only small side chains are tolerable. These results further elucidate the factors governing the region of the β-bulge structure that includes Gly(17), illuminating why glycine is conserved in all mammalian α-defensins.
- The 1st Affiliated Hospital, Xi'an Jiaotong University School of Medicine, China.
Organizational Affiliation: