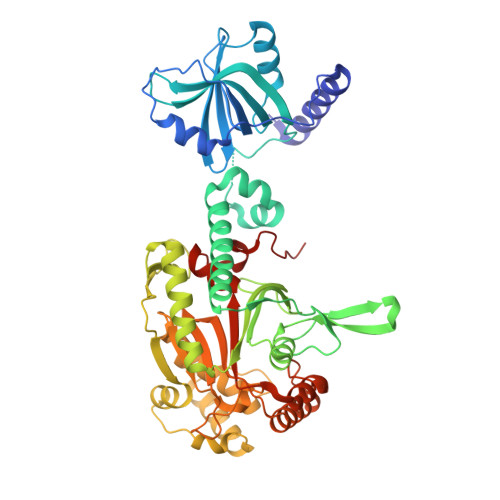
Structural Switch of Lysyl-tRNA Synthetase between Translation and Transcription.
Ofir-Birin, Y., Fang, P., Bennett, S.P., Zhang, H.M., Wang, J., Rachmin, I., Shapiro, R., Song, J., Dagan, A., Pozo, J., Kim, S., Marshall, A.G., Schimmel, P., Yang, X.L., Nechushtan, H., Razin, E., Guo, M.(2013) Mol Cell 49: 30-42
- PubMed: 23159739
- DOI: https://doi.org/10.1016/j.molcel.2012.10.010
- Primary Citation of Related Structures:
4DPG - PubMed Abstract:
Lysyl-tRNA synthetase (LysRS), a component of the translation apparatus, is released from the cytoplasmic multi-tRNA synthetase complex (MSC) to activate the transcription factor MITF in stimulated mast cells through undefined mechanisms. Here we show that Ser207 phosphorylation provokes a new conformer of LysRS that inactivates its translational function but activates its transcriptional function. The crystal structure of an MSC subcomplex established that LysRS is held in the MSC by binding to the N terminus of the scaffold protein p38/AIMP2. Phosphorylation-created steric clashes at the LysRS domain interface disrupt its binding grooves for p38/AIMP2, releasing LysRS and provoking its nuclear translocation. This alteration also exposes the C-terminal domain of LysRS to bind to MITF and triggers LysRS-directed production of the second messenger Ap(4)A that activates MITF. Thus our results establish that a single conformational change triggered by phosphorylation leads to multiple effects driving an exclusive switch of LysRS function from translation to transcription.
- Department of Biochemistry and Molecular Biology, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel.
Organizational Affiliation: