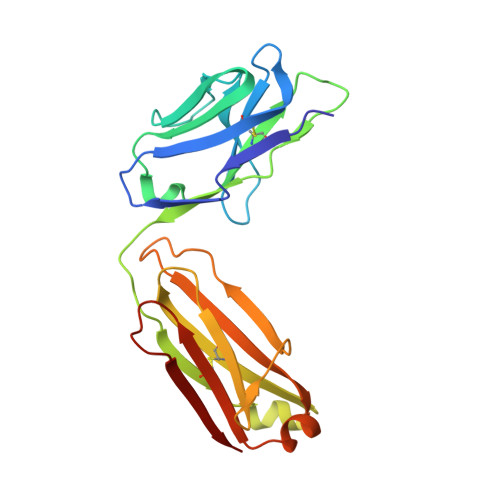
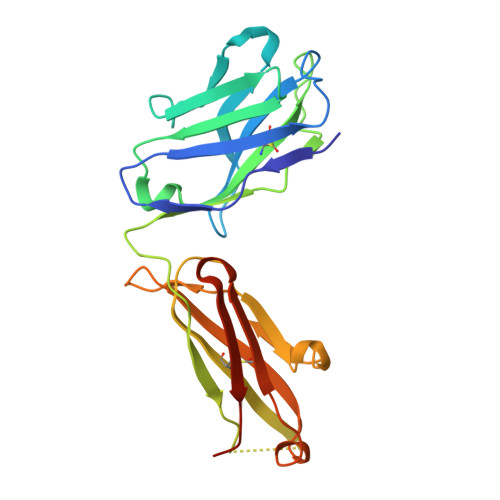
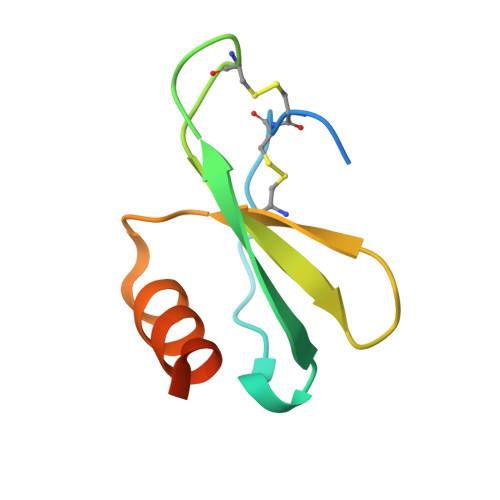
Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Obmolova, G., Teplyakov, A., Malia, T.J., Grygiel, T.L., Sweet, R., Snyder, L.A., Gilliland, G.L.(2012) Mol Immunol 51: 227-233
- PubMed: 22487721
- DOI: https://doi.org/10.1016/j.molimm.2012.03.022
- Primary Citation of Related Structures:
4DN3, 4DN4 - PubMed Abstract:
Human CC chemokine ligand 2 (CCL2), also known as monocyte chemoattractant protein-1 (MCP-1), is a member of the β chemokine family whose actions are mediated through the G-protein-coupled receptor CCR2. Binding of CCL2 to its receptor CCR2 triggers calcium mobilization and chemotaxis. CCL2 is implicated in the pathogenesis of certain inflammatory diseases and cancer. CNTO 888, a neutralizing human anti-CCL2 antibody, was derived by antibody phage display. The antibody binds human CCL2 with high affinity (K(D)=22 pM) and inhibits CCL2 binding to its receptor. The crystal structure of the CNTO 888 Fab alone and in complex with the monomeric form of CCL2 (P8A variant) was determined at 2.6 Å and 2.8 Å resolution, respectively. CNTO 888 recognizes a conformational epitope encompassing residues 18-24 and 45-51 that overlaps the mapped receptor binding site. The epitope of CNTO 888 does not overlap with the dimerization site of CCL2, and thus its inhibitory activity is not expected to result from interference with the oligomeric state of CCL2. Comparison of the X-ray-determined epitopes of CNTO 888 and another CCL2-neutralizing antibody, 11K2, provides insight into the molecular basis of antibody selectivity and functional inhibition.
- Discovery Research, Janssen R&D LLC, 145 King of Prussia Road, Radnor, PA 19087, USA.
Organizational Affiliation: