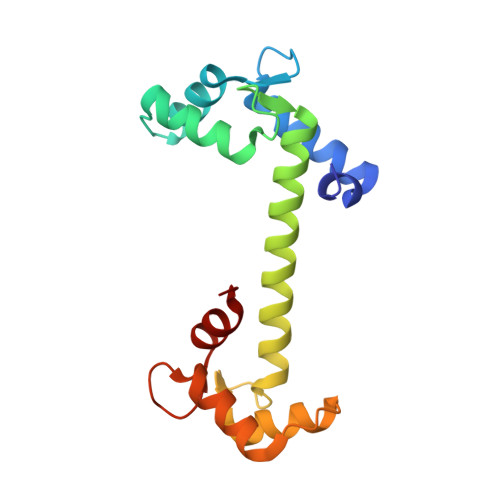
Crystallographic basis for calcium regulation of sodium channels.
Sarhan, M.F., Tung, C.C., Van Petegem, F., Ahern, C.A.(2012) Proc Natl Acad Sci U S A 109: 3558-3563
- PubMed: 22331908
- DOI: https://doi.org/10.1073/pnas.1114748109
- Primary Citation of Related Structures:
4DJC - PubMed Abstract:
Voltage-gated sodium channels underlie the rapid regenerative upstroke of action potentials and are modulated by cytoplasmic calcium ions through a poorly understood mechanism. We describe the 1.35 Å crystal structure of Ca(2+)-bound calmodulin (Ca(2+)/CaM) in complex with the inactivation gate (DIII-IV linker) of the cardiac sodium channel (Na(V)1.5). The complex harbors the positions of five disease mutations involved with long Q-T type 3 and Brugada syndromes. In conjunction with isothermal titration calorimetry, we identify unique inactivation-gate mutations that enhance or diminish Ca(2+)/CaM binding, which, in turn, sensitize or abolish Ca(2+) regulation of full-length channels in electrophysiological experiments. Additional biochemical experiments support a model whereby a single Ca(2+)/CaM bridges the C-terminal IQ motif to the DIII-IV linker via individual N and C lobes, respectively. The data suggest that Ca(2+)/CaM destabilizes binding of the inactivation gate to its receptor, thus biasing inactivation toward more depolarized potentials.
- Department of Anesthesiology, Pharmacology and Therapeutics, Life Sciences Institute, University of British Columbia, Vancouver, BC, Canada, V6T 1Z3.
Organizational Affiliation: