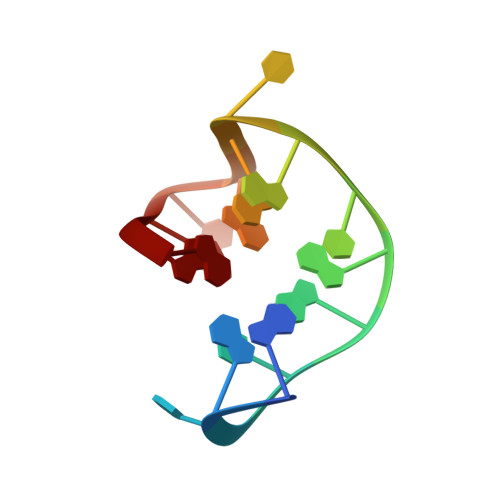
High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Russo Krauss, I., Merlino, A., Randazzo, A., Novellino, E., Mazzarella, L., Sica, F.(2012) Nucleic Acids Res 40: 8119-8128
- PubMed: 22669903
- DOI: https://doi.org/10.1093/nar/gks512
- Primary Citation of Related Structures:
4DIH, 4DII - PubMed Abstract:
The G-quadruplex architecture is a peculiar structure adopted by guanine-rich oligonucleotidic sequences, and, in particular, by several aptamers, including the thrombin-binding aptamer (TBA) that has the highest inhibitory activity against human α-thrombin. A crucial role in determining structure, stability and biological properties of G-quadruplexes is played by ions. In the case of TBA, K(+) ions cause an enhancement of the aptamer clotting inhibitory activity. A detailed picture of the interactions of TBA with the protein and with the ions is still lacking, despite the importance of this aptamer in biomedical field for detection and inhibition of α-thrombin. Here, we fill this gap by presenting a high-resolution crystallographic structural characterization of the thrombin-TBA complex formed in the presence of Na(+) or K(+) and a circular dichroism study of the structural stability of the aptamer both free and complexed with α-thrombin, in the presence of the two ionic species. The results indicate that the different effects exerted by Na(+) and K(+) on the inhibitory activity of TBA are related to a subtle perturbation of a few key interactions at the protein-aptamer interface. The present data, in combination with those previously obtained on the complex between α-thrombin and a modified aptamer, may allow the design of new TBA variants with a pharmacological performance enhancement.
- Dipartimento di Scienze Chimiche, Università di Napoli Federico II, Via Cintia, I-80126 Napoli, Italia.
Organizational Affiliation: