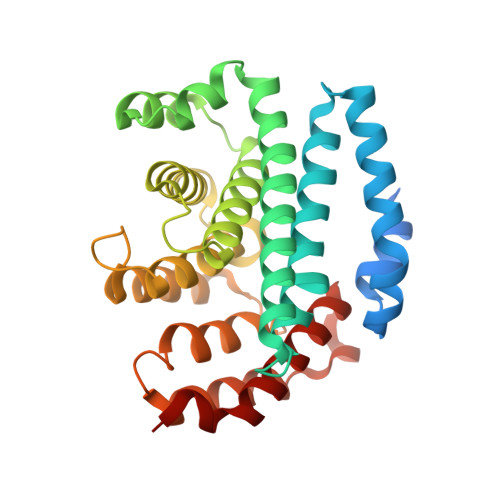
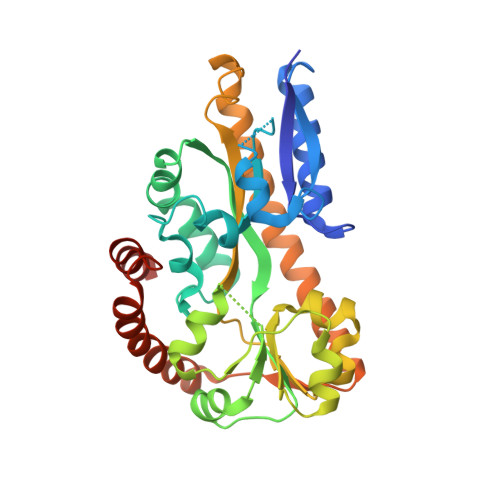
Structural and Thermodynamic Characterization of the Interaction between Two Periplasmic Treponema pallidum Lipoproteins that are Components of a TPR-Protein-Associated TRAP Transporter (TPAT).
Brautigam, C.A., Deka, R.K., Schuck, P., Tomchick, D.R., Norgard, M.V.(2012) J Mol Biology 420: 70-86
- PubMed: 22504226
- DOI: https://doi.org/10.1016/j.jmb.2012.04.001
- Primary Citation of Related Structures:
4DI3, 4DI4 - PubMed Abstract:
Tripartite ATP-independent periplasmic transporters (TRAP-Ts) are bacterial transport systems that have been implicated in the import of small molecules into the cytoplasm. A newly discovered subfamily of TRAP-Ts [tetratricopeptide repeat-protein associated TRAP transporters (TPATs)] has four components. Three are common to both TRAP-Ts and TPATs: the P component, a ligand-binding protein, and a transmembrane symporter apparatus comprising the M and Q components (M and Q are sometimes fused to form a single polypeptide). TPATs are distinguished from TRAP-Ts by the presence of a unique protein called the "T component". In Treponema pallidum, this protein (TatT) is a water-soluble trimer whose protomers are each perforated by a pore. Its respective P component (TatP(T)) interacts with the TatT in vitro and in vivo. In this work, we further characterized this interaction. Co-crystal structures of two complexes between the two proteins confirm that up to three monomers of TatP(T) can bind to the TatT trimer. A putative ligand-binding cleft of TatP(T) aligns with the pore of TatT, strongly suggesting ligand transfer between T and P(T). We used a combination of site-directed mutagenesis and analytical ultracentrifugation to derive thermodynamic parameters for the interactions. These observations confirm that the observed crystallographic interface is recapitulated in solution. These results prompt a hypothesis of the molecular mechanism(s) of hydrophobic ligand transport by the TPATs.
- Department of Biochemistry, The University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: