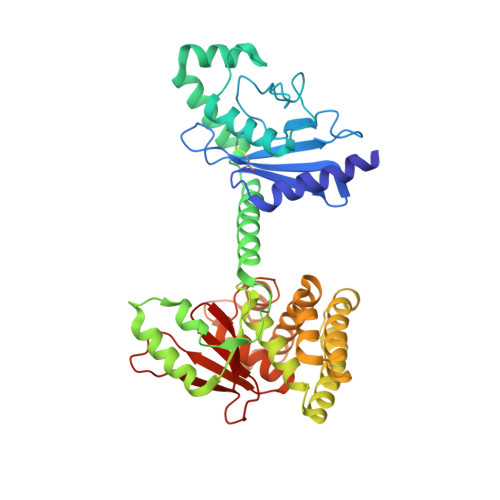
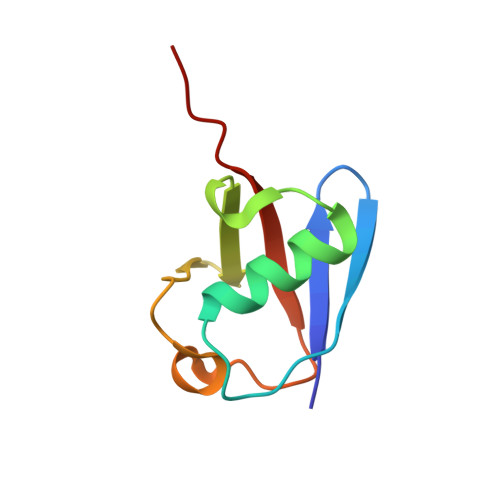
OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function.
Juang, Y.C., Landry, M.C., Sanches, M., Vittal, V., Leung, C.C., Ceccarelli, D.F., Mateo, A.R., Pruneda, J.N., Mao, D.Y., Szilard, R.K., Orlicky, S., Munro, M., Brzovic, P.S., Klevit, R.E., Sicheri, F., Durocher, D.(2012) Mol Cell 45: 384-397
- PubMed: 22325355
- DOI: https://doi.org/10.1016/j.molcel.2012.01.011
- Primary Citation of Related Structures:
4DDG, 4DDI - PubMed Abstract:
Ubiquitylation entails the concerted action of E1, E2, and E3 enzymes. We recently reported that OTUB1, a deubiquitylase, inhibits the DNA damage response independently of its isopeptidase activity. OTUB1 does so by blocking ubiquitin transfer by UBC13, the cognate E2 enzyme for RNF168. OTUB1 also inhibits E2s of the UBE2D and UBE2E families. Here we elucidate the structural mechanism by which OTUB1 binds E2s to inhibit ubiquitin transfer. OTUB1 recognizes ubiquitin-charged E2s through contacts with both donor ubiquitin and the E2 enzyme. Surprisingly, free ubiquitin associates with the canonical distal ubiquitin-binding site on OTUB1 to promote formation of the inhibited E2 complex. Lys48 of donor ubiquitin lies near the OTUB1 catalytic site and the C terminus of free ubiquitin, a configuration that mimics the products of Lys48-linked ubiquitin chain cleavage. OTUB1 therefore co-opts Lys48-linked ubiquitin chain recognition to suppress ubiquitin conjugation and the DNA damage response.
- Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, ON M5G 1X5, Canada.
Organizational Affiliation: