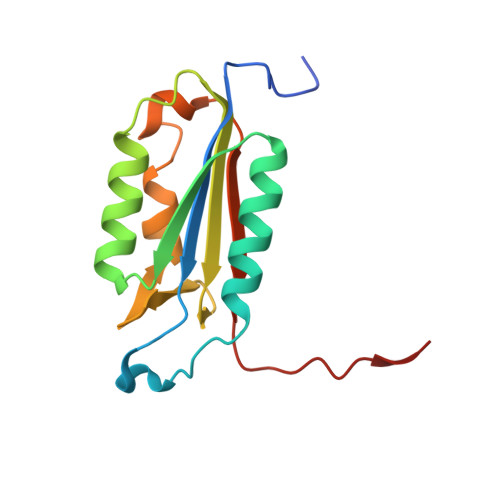
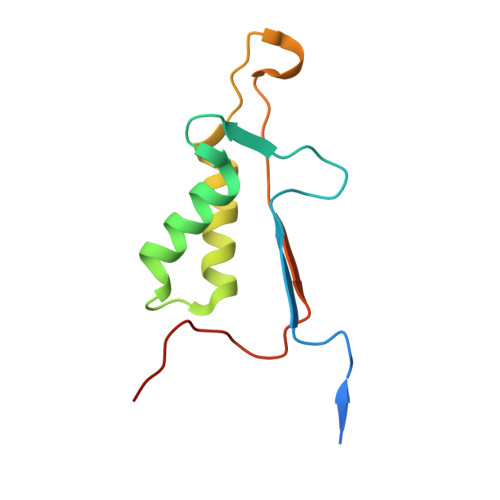
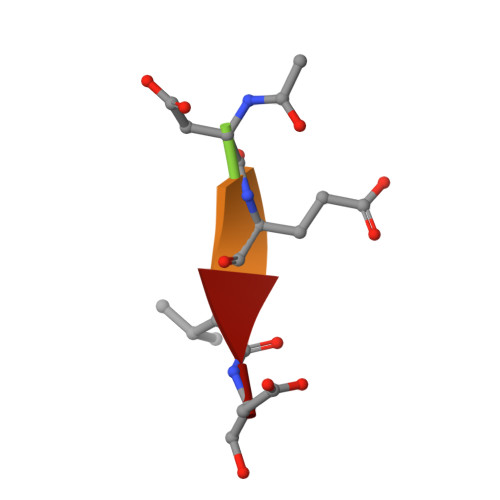
Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Kang, H.J., Lee, Y.M., Jeong, M.S., Kim, M., Bae, K.H., Kim, S.J., Chung, S.J.(2012) Biosci Rep 32: 305-313
- PubMed: 22304005
- DOI: https://doi.org/10.1042/BSR20120009
- Primary Citation of Related Structures:
4DCJ, 4DCO, 4DCP - PubMed Abstract:
Various apoptotic signals can activate caspases 3 and 7 by triggering the L2 loop cleavage of their proenzymes. These two enzymes have highly similar structures and functions, and serve as apoptotic executioners. The structures of caspase 7 and procaspase 7 differ significantly in the conformation of the loops constituting the active site, indicating that the enzyme undergoes a large structural change during activation. To define the role of the leucine residue on the L2 loop, which shows the largest movement during enzyme activation but has not yet been studied, Leu168 of caspase 3 and Leu191 of caspase 7 were mutated. Kinetic analysis indicated that the mutation of the leucine residues sometimes improved the Km but also greatly decreased the kcat, resulting in an overall decrease in enzyme activity. The tryptophan fluorescence change at excitation/emission = 280/350 nm upon L2-L2' loop cleavage was found to be higher in catalytically active mutants, including the corresponding wild-type caspase, than in the inactive mutants. The crystal structures of the caspase 3 mutants were solved and compared with that of wild-type. Significant alterations in the conformations of the L1 and L4 loops were found. These results indicate that the leucine residue on the L2 loop has an important role in maintaining the catalytic activity of caspases 3 and 7.
- BioNanotechnology Research Center, KRIBB, Yuseong, Daejeon, 305-806, Korea.
Organizational Affiliation: