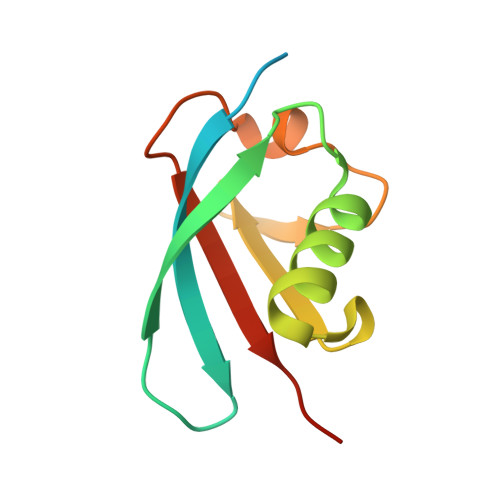
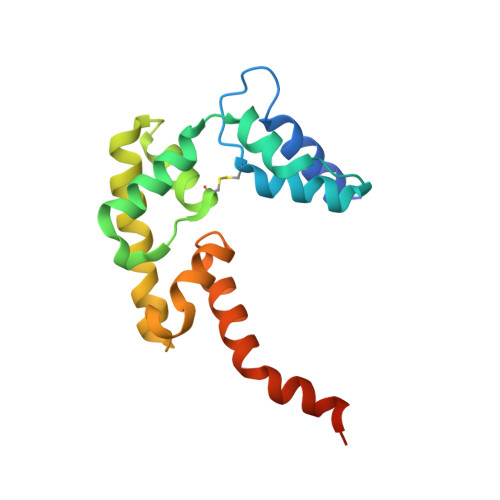
A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex
Yagi, H., Ishimoto, K., Hiromoto, T., Fujita, H., Mizushima, T., Uekusa, Y., Yagi-Utsumi, M., Kurimoto, E., Noda, M., Uchiyama, S., Tokunaga, F., Iwai, K., Kato, K.(2012) EMBO Rep 13: 462-468
- PubMed: 22430200
- DOI: https://doi.org/10.1038/embor.2012.24
- Primary Citation of Related Structures:
4DBG - PubMed Abstract:
HOIL-1L and its binding partner HOIP are essential components of the E3-ligase complex that generates linear ubiquitin (Ub) chains, which are critical regulators of NF-κB activation. Using crystallographic and mutational approaches, we characterize the unexpected structural basis for the specific interaction between the Ub-like domain (UBL) of HOIL-1L and the Ub-associated domain (UBA) of HOIP. Our data indicate the functional significance of this non-canonical mode of UBA-UBL interaction in E3 complex formation and subsequent NF-κB activation. This study highlights the versatility and specificity of protein-protein interactions involving Ub/UBLs and their cognate proteins.
- Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan.
Organizational Affiliation: