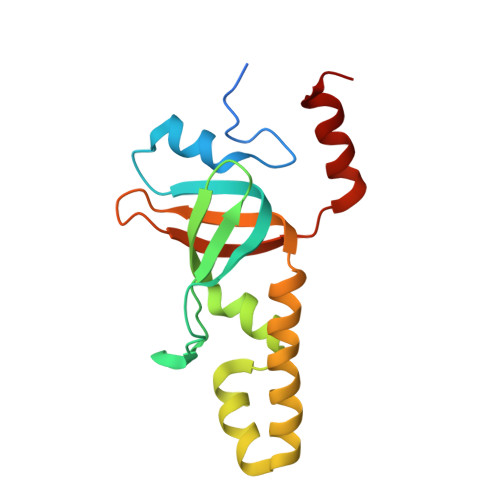
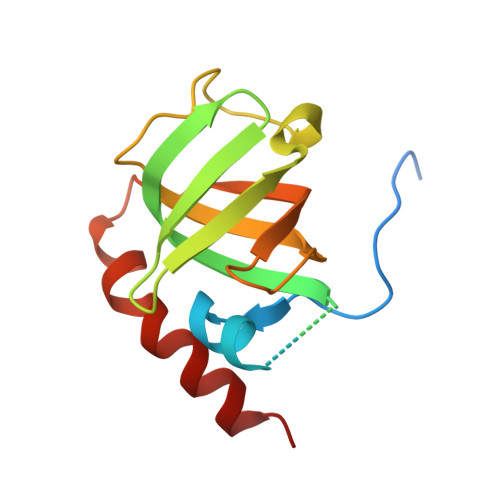
Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome.
Hoadley, K.A., Xue, Y., Ling, C., Takata, M., Wang, W., Keck, J.L.(2012) Proc Natl Acad Sci U S A 109: 4437-4442
- PubMed: 22392978
- DOI: https://doi.org/10.1073/pnas.1117279109
- Primary Citation of Related Structures:
4DAY - PubMed Abstract:
The RMI subcomplex (RMI1/RMI2) functions with the BLM helicase and topoisomerase IIIα in a complex called the "dissolvasome," which separates double-Holliday junction DNA structures that can arise during DNA repair. This activity suppresses potentially harmful sister chromatid exchange (SCE) events in wild-type cells but not in cells derived from Bloom syndrome patients with inactivating BLM mutations. The RMI subcomplex also associates with FANCM, a component of the Fanconi anemia (FA) core complex that is important for repair of stalled DNA replication forks. The RMI/FANCM interface appears to help coordinate dissolvasome and FA core complex activities, but its precise role remains poorly understood. Here, we define the structure of the RMI/FANCM interface and investigate its roles in coordinating cellular DNA-repair activities. The X-ray crystal structure of the RMI core complex bound to a well-conserved peptide from FANCM shows that FANCM binds to both RMI proteins through a hydrophobic "knobs-into-holes" packing arrangement. The RMI/FANCM interface is shown to be critical for interaction between the components of the dissolvasome and the FA core complex. FANCM variants that substitute alanine for key interface residues strongly destabilize the complex in solution and lead to increased SCE levels in cells that are similar to those observed in blm- or fancm-deficient cells. This study provides a molecular view of the RMI/FANCM complex and highlights a key interface utilized in coordinating the activities of two critical eukaryotic DNA-damage repair machines.
- Department of Biomolecular Chemistry, 420 Henry Mall, University of Wisconsin School of Medicine and Public Health, Madison, WI 53706, USA.
Organizational Affiliation: