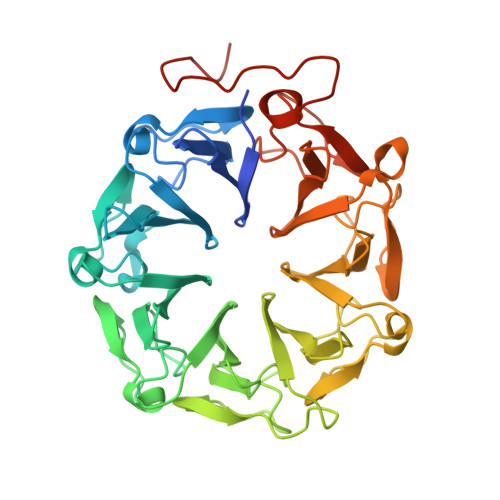
Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Christie, J.M., Arvai, A.S., Baxter, K.J., Heilmann, M., Pratt, A.J., O'Hara, A., Kelly, S.M., Hothorn, M., Smith, B.O., Hitomi, K., Jenkins, G.I., Getzoff, E.D.(2012) Science 335: 1492-1496
- PubMed: 22323738
- DOI: https://doi.org/10.1126/science.1218091
- Primary Citation of Related Structures:
4D9S - PubMed Abstract:
The recently identified plant photoreceptor UVR8 (UV RESISTANCE LOCUS 8) triggers regulatory changes in gene expression in response to ultraviolet-B (UV-B) light through an unknown mechanism. Here, crystallographic and solution structures of the UVR8 homodimer, together with mutagenesis and far-UV circular dichroism spectroscopy, reveal its mechanisms for UV-B perception and signal transduction. β-propeller subunits form a remarkable, tryptophan-dominated, dimer interface stitched together by a complex salt-bridge network. Salt-bridging arginines flank the excitonically coupled cross-dimer tryptophan "pyramid" responsible for UV-B sensing. Photoreception reversibly disrupts salt bridges, triggering dimer dissociation and signal initiation. Mutation of a single tryptophan to phenylalanine retunes the photoreceptor to detect UV-C wavelengths. Our analyses establish how UVR8 functions as a photoreceptor without a prosthetic chromophore to promote plant development and survival in sunlight.
- Institute of Molecular, Cell and Systems Biology, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, UK.
Organizational Affiliation: