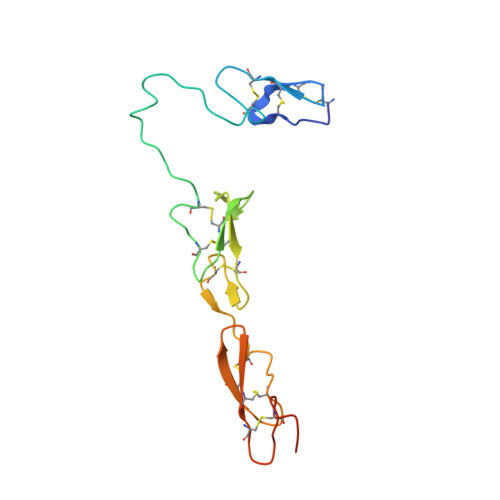
The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Schurpf, T., Chen, Q., Liu, J.H., Wang, R., Springer, T.A., Wang, J.H.(2012) FASEB J 26: 3412-3420
- PubMed: 22601780
- DOI: https://doi.org/10.1096/fj.11-202036
- Primary Citation of Related Structures:
4D90 - PubMed Abstract:
Developmental endothelial cell locus-1 (Del-1) glycoprotein is secreted by endothelial cells and a subset of macrophages. Del-1 plays a regulatory role in vascular remodeling and functions in innate immunity through interaction with integrin α(V)β(3). Del-1 contains 3 epidermal growth factor (EGF)-like repeats and 2 discoidin-like domains. An Arg-Gly-Asp (RGD) motif in the second EGF domain (EGF2) mediates adhesion by endothelial cells and phagocytes. We report the crystal structure of its 3 EGF domains. The RGD motif of EGF2 forms a type II' β turn at the tip of a long protruding loop, dubbed the RGD finger. Whereas EGF2 and EGF3 constitute a rigid rod via an interdomain calcium ion binding site, the long linker between EGF1 and EGF2 lends considerable flexibility to EGF1. Two unique O-linked glycans and 1 N-linked glycan locate to the opposite side of EGF2 from the RGD motif. These structural features favor integrin binding of the RGD finger. Mutagenesis data confirm the importance of having the RGD motif at the tip of the RGD finger. A database search for EGF domain sequences shows that this RGD finger is likely an evolutionary insertion and unique to the EGF domain of Del-1 and its homologue milk fat globule-EGF 8.
- Immune Disease Institute, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: