An Unprecedented Nadph Domain Conformation in Lysine Monooxygenase Nbtg Provides Insights Into Uncoupling of Oxygen Consumption from Substrate Hydroxylation.
Binda, C., Robinson, R.M., Martin Del Campo, J.S., Keul, N.D., Rodriguez, P.J., Robinson, H.H., Mattevi, A., Sobrado, P.(2015) J Biological Chem 290: 12676
- PubMed: 25802330
- DOI: https://doi.org/10.1074/jbc.M114.629485
- Primary Citation of Related Structures:
4D7E - PubMed Abstract:
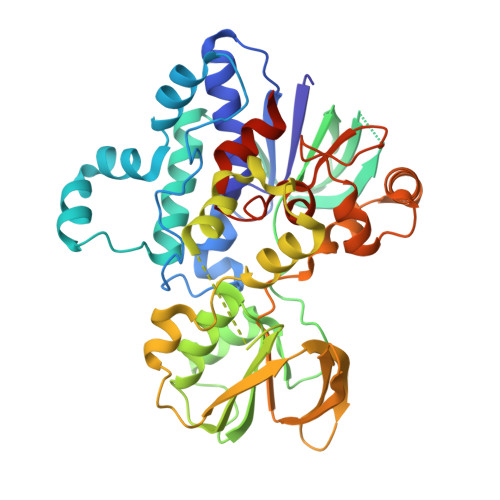
N-Hydroxylating monooxygenases are involved in the biosynthesis of iron-chelating hydroxamate-containing siderophores that play a role in microbial virulence. These flavoenzymes catalyze the NADPH- and oxygen-dependent hydroxylation of amines such as those found on the side chains of lysine and ornithine. In this work we report the biochemical and structural characterization of Nocardia farcinica Lys monooxygenase (NbtG), which has similar biochemical properties to mycobacterial homologs. NbtG is also active on d-Lys, although it binds l-Lys with a higher affinity. Differently from the ornithine monooxygenases PvdA, SidA, and KtzI, NbtG can use both NADH and NADPH and is highly uncoupled, producing more superoxide and hydrogen peroxide than hydroxylated Lys. The crystal structure of NbtG solved at 2.4 Å resolution revealed an unexpected protein conformation with a 30° rotation of the NAD(P)H domain with respect to the flavin adenine dinucleotide (FAD) domain that precludes binding of the nicotinamide cofactor. This "occluded" structure may explain the biochemical properties of NbtG, specifically with regard to the substantial uncoupling and limited stabilization of the C4a-hydroperoxyflavin intermediate. Biological implications of these findings are discussed.
- From the Department of Biology and Biotechnology, University of Pavia, Pavia 27100, Italy.
Organizational Affiliation: