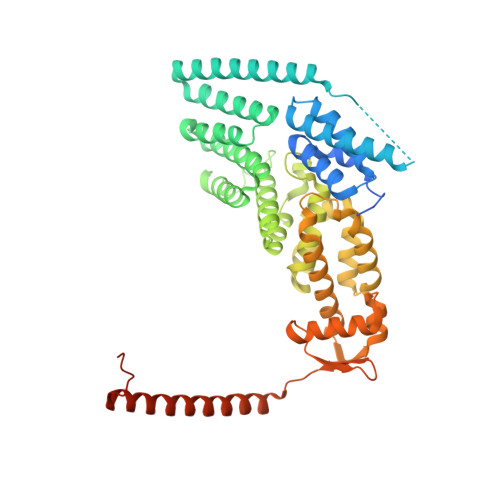
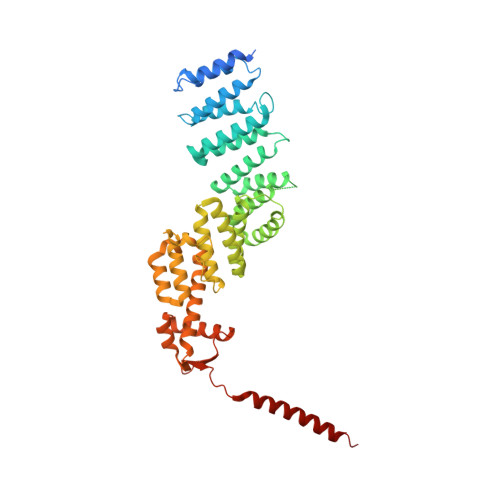
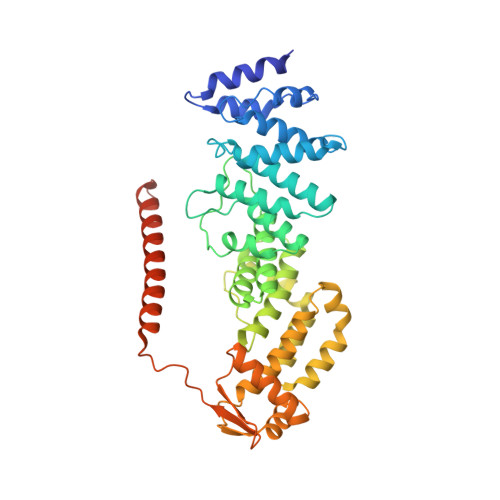
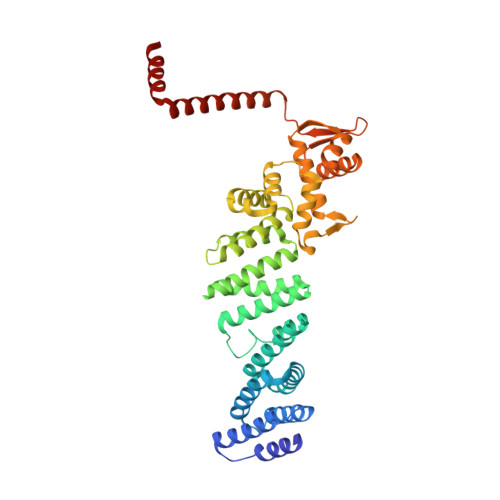
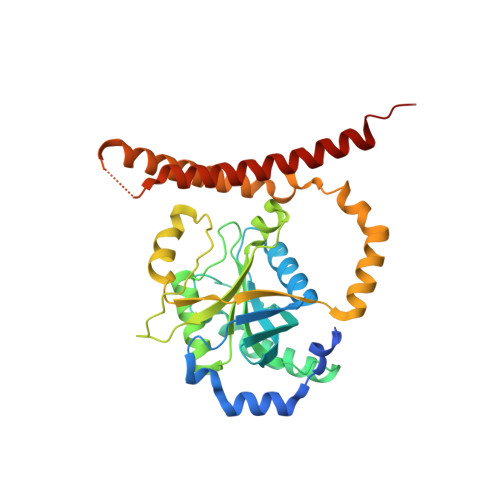
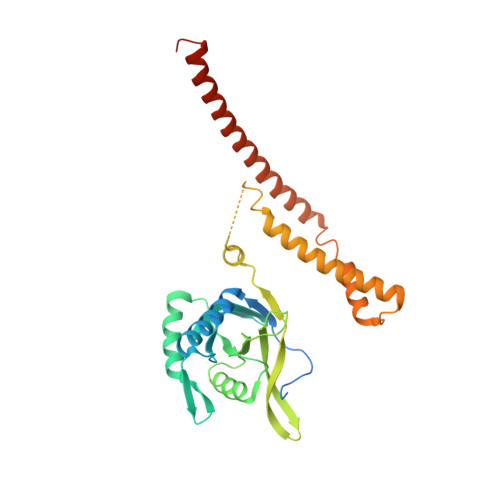
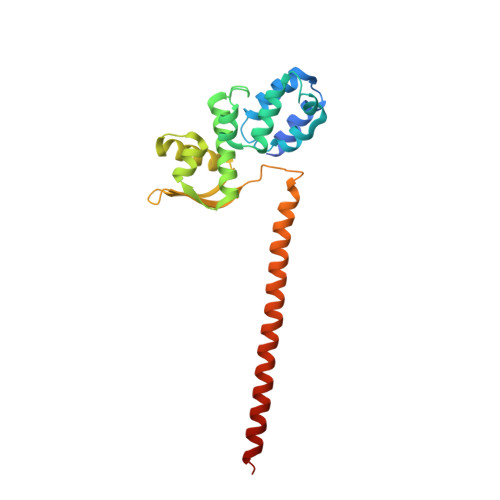
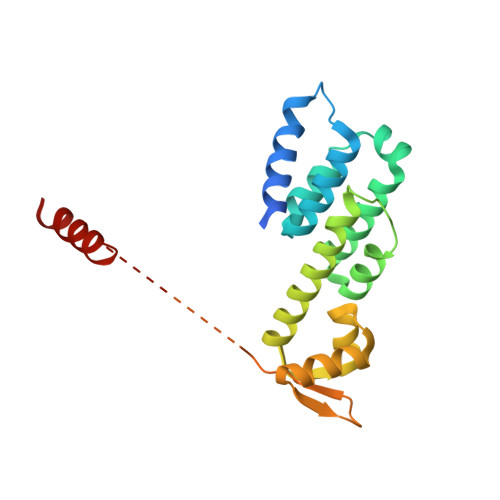
Crystal Structure of the Human Cop9 Signalosome
Lingaraju, G.M., Bunker, R.D., Cavadini, S., Hess, D., Hassiepen, U., Renatus, M., Fischer, E.S., Thoma, N.H.(2014) Nature 512: 161
- PubMed: 25043011
- DOI: https://doi.org/10.1038/nature13566
- Primary Citation of Related Structures:
4D0P, 4D10, 4D18 - PubMed Abstract:
Ubiquitination is a crucial cellular signalling process, and is controlled on multiple levels. Cullin-RING E3 ubiquitin ligases (CRLs) are regulated by the eight-subunit COP9 signalosome (CSN). CSN inactivates CRLs by removing their covalently attached activator, NEDD8. NEDD8 cleavage by CSN is catalysed by CSN5, a Zn(2+)-dependent isopeptidase that is inactive in isolation. Here we present the crystal structure of the entire ∼350-kDa human CSN holoenzyme at 3.8 Å resolution, detailing the molecular architecture of the complex. CSN has two organizational centres: a horseshoe-shaped ring created by its six proteasome lid-CSN-initiation factor 3 (PCI) domain proteins, and a large bundle formed by the carboxy-terminal α-helices of every subunit. CSN5 and its dimerization partner, CSN6, are intricately embedded at the core of the helical bundle. In the substrate-free holoenzyme, CSN5 is autoinhibited, which precludes access to the active site. We find that neddylated CRL binding to CSN is sensed by CSN4, and communicated to CSN5 with the assistance of CSN6, resulting in activation of the deneddylase.
- 1] Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, 4058 Basel, Switzerland [2] University of Basel, Petersplatz 10, 4003 Basel, Switzerland [3].
Organizational Affiliation: