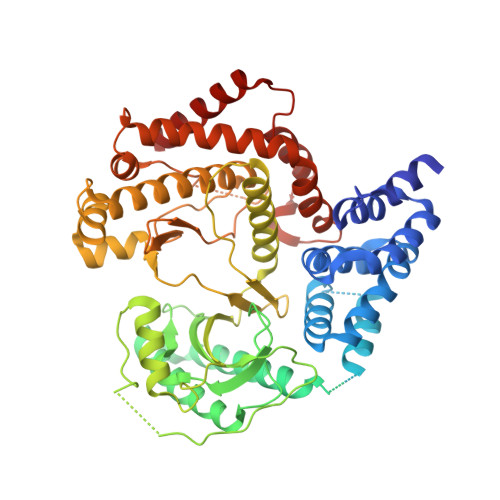
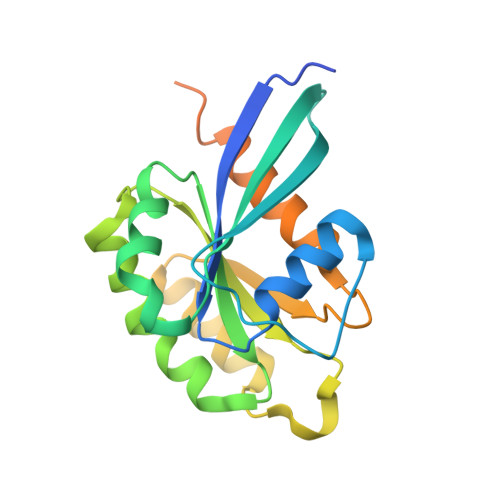
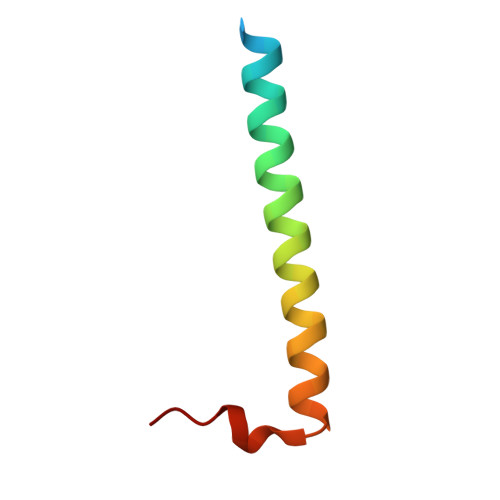
Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Burke, J.E., Inglis, A.J., Perisic, O., Masson, G.R., Mclaughlin, S.H., Rutaganira, F., Shokat, K.M., Williams, R.L.(2014) Science 344: 1035
- PubMed: 24876499
- DOI: https://doi.org/10.1126/science.1253397
- Primary Citation of Related Structures:
4D0L, 4D0M - PubMed Abstract:
Phosphatidylinositol 4-kinases (PI4Ks) and small guanosine triphosphatases (GTPases) are essential for processes that require expansion and remodeling of phosphatidylinositol 4-phosphate (PI4P)-containing membranes, including cytokinesis, intracellular development of malarial pathogens, and replication of a wide range of RNA viruses. However, the structural basis for coordination of PI4K, GTPases, and their effectors is unknown. Here, we describe structures of PI4Kβ (PI4KIIIβ) bound to the small GTPase Rab11a without and with the Rab11 effector protein FIP3. The Rab11-PI4KIIIβ interface is distinct compared with known structures of Rab complexes and does not involve switch regions used by GTPase effectors. Our data provide a mechanism for how PI4KIIIβ coordinates Rab11 and its effectors on PI4P-enriched membranes and also provide strategies for the design of specific inhibitors that could potentially target plasmodial PI4KIIIβ to combat malaria.
- Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge CB2 0QH, UK. jeburke@uvic.ca rlw@mrc-lmb.cam.ac.uk.
Organizational Affiliation: