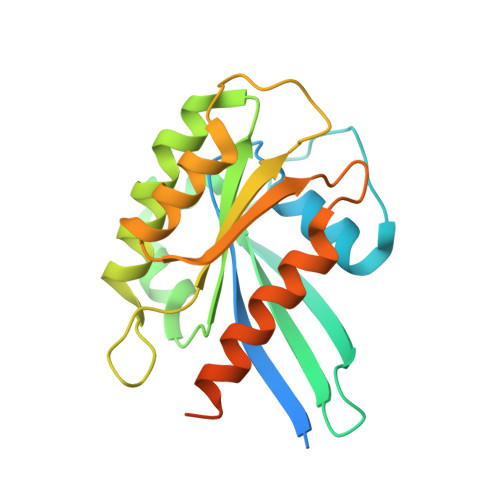
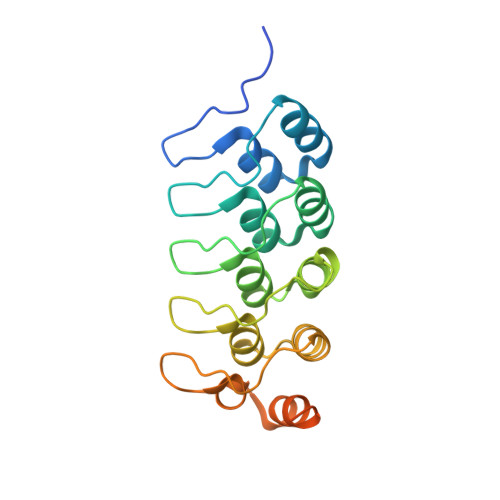
Varp is Recruited on to Endosomes by Direct Interaction with Retromer, Where Together They Function in Export to the Cell Surface.
Hesketh, G.G., Perez-Dorado, I., Jackson, L.P., Wartosch, L., Schefer, I.B., Gray, S.R., Mccoy, A.J., Zeldin, O.B., Garman, E.F., Harbour, M.E., Evans, P.R., Seaman, M.N., Luzio, J.P., Owen, D.J.(2014) Dev Cell 29: 591
- PubMed: 24856514
- DOI: https://doi.org/10.1016/j.devcel.2014.04.010
- Primary Citation of Related Structures:
4CYM, 4CZ2 - PubMed Abstract:
VARP is a Rab32/38 effector that also binds to the endosomal/lysosomal R-SNARE VAMP7. VARP binding regulates VAMP7 participation in SNARE complex formation and can therefore influence VAMP7-mediated membrane fusion events. Mutant versions of VARP that cannot bind Rab32:GTP, designed on the basis of the VARP ankyrin repeat/Rab32:GTP complex structure described here, unexpectedly retain endosomal localization, showing that VARP recruitment is not dependent on Rab32 binding. We show that recruitment of VARP to the endosomal membrane is mediated by its direct interaction with VPS29, a subunit of the retromer complex, which is involved in trafficking from endosomes to the TGN and the cell surface. Transport of GLUT1 from endosomes to the cell surface requires VARP, VPS29, and VAMP7 and depends on the direct interaction between VPS29 and VARP. Finally, we propose that endocytic cycling of VAMP7 depends on its interaction with VARP and, consequently, also on retromer.
- Cambridge Institute for Medical Research and Department of Clinical Biochemistry, University of Cambridge, Wellcome Trust/MRC Building, Cambridge Biomedical Campus, Hills Road, Cambridge CB2 0XY, UK.
Organizational Affiliation: