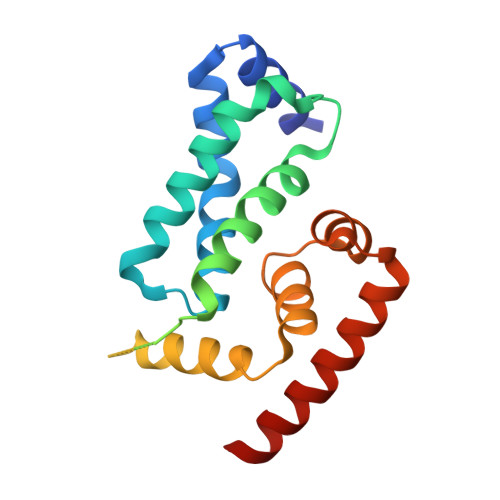
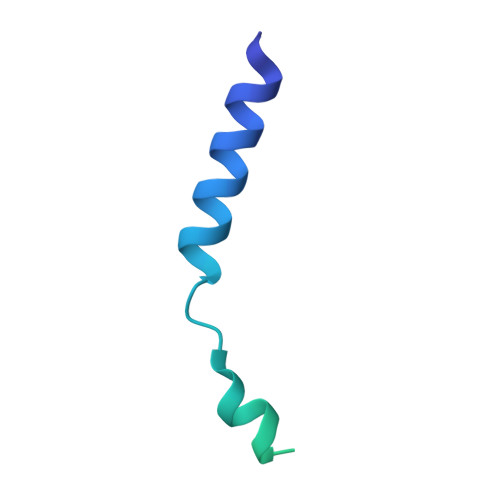
The Crystal Structure of the Anti-Sigma Factor Cnry in Complex with the Sigma Factor Cnrh Shows a New Structural Class of Anti- Sigma Factors Targeting Extracytoplasmic-Function Sigma Factors.
Maillard, A.P., Girard, E., Ziani, W., Petit-Hartlein, I., Kahn, R., Coves, J.(2014) J Mol Biology 426: 2313
- PubMed: 24727125
- DOI: https://doi.org/10.1016/j.jmb.2014.04.003
- Primary Citation of Related Structures:
4CXF - PubMed Abstract:
Gene expression in bacteria is regulated at the level of transcription initiation, a process driven by σ factors. The regulation of σ factor activity proceeds from the regulation of their cytoplasmic availability, which relies on specific inhibitory proteins called anti-σ factors. With anti-σ factors regulating their availability according to diverse cues, extracytoplasmic function σ factors (σ(ECF)) form a major signal transduction system in bacteria. Here, structure:function relationships have been characterized in an emerging class of minimal-size transmembrane anti-σ factors, using CnrY from Cupriavidus metallidurans CH34 as a model. This study reports the 1.75-Å-resolution structure of CnrY cytosolic domain in complex with CnrH, its cognate σ(ECF), and identifies a small hydrophobic knob in CnrY as the major determinant of this interaction in vivo. Unsuspected structural similarity with the molecular switch regulating the general stress response in α-proteobacteria unravels a new class of anti-σ factors targeting σ(ECF). Members of this class carry out their function via a 30-residue stretch that displays helical propensity but no canonical structure on its own.
- Université Grenoble Alpes, Commissariat à l'Energie Atomique et aux Energies Alternatives, Centre National de la Recherche Scientifique, and Institut de Biologie Structurale, F-38000 Grenoble, France. Electronic address: antoine.maillard@ibs.fr.
Organizational Affiliation: