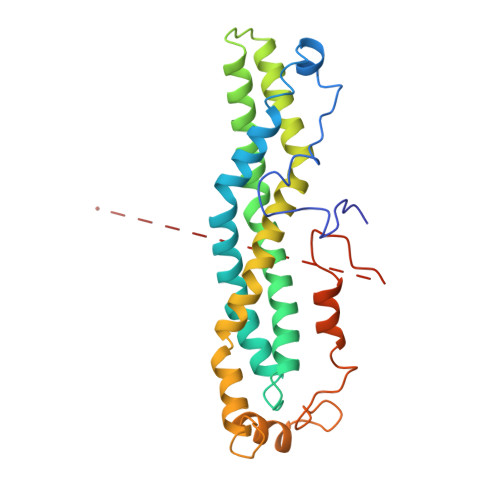
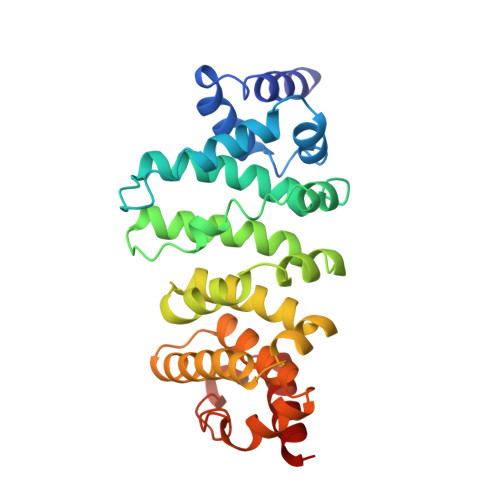
A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Chen, Y., Boland, A., Kuzuoglu-Ozturk, D., Bawankar, P., Loh, B., Chang, C.T., Weichenrieder, O., Izaurralde, E.(2014) Mol Cell 54: 737
- PubMed: 24768540
- DOI: https://doi.org/10.1016/j.molcel.2014.03.034
- Primary Citation of Related Structures:
4CRU, 4CRV, 4CRW - PubMed Abstract:
CCR4-NOT is a major effector complex in miRNA-mediated gene silencing. It is recruited to miRNA targets through interactions with tryptophan (W)-containing motifs in TNRC6/GW182 proteins and is required for both translational repression and degradation of miRNA targets. Here, we elucidate the structural basis for the repressive activity of CCR4-NOT and its interaction with TNRC6/GW182s. We show that the conserved CNOT9 subunit attaches to a domain of unknown function (DUF3819) in the CNOT1 scaffold. The resulting complex provides binding sites for TNRC6/GW182, and its crystal structure reveals tandem W-binding pockets located in CNOT9. We further show that the CNOT1 MIF4G domain interacts with the C-terminal RecA domain of DDX6, a translational repressor and decapping activator. The crystal structure of this complex demonstrates striking similarity to the eIF4G-eIF4A complex. Together, our data provide the missing physical links in a molecular pathway that connects miRNA target recognition with translational repression, deadenylation, and decapping.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Spemannstrasse 35, 72076 Tübingen, Germany.
Organizational Affiliation: