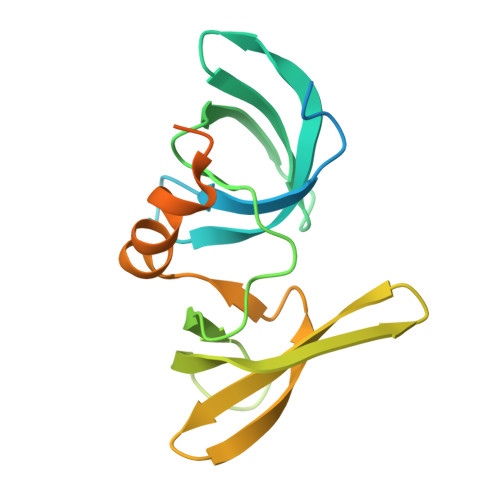
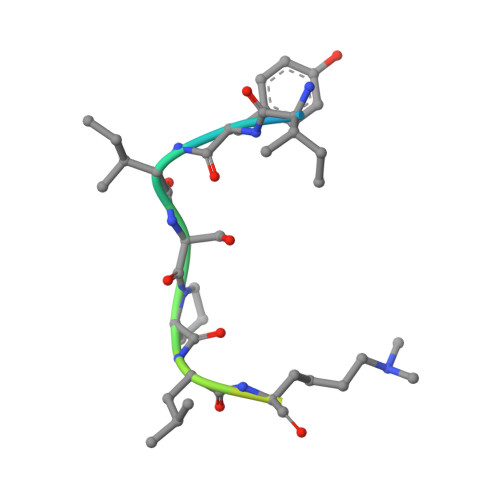
Lysine Methylation-Dependent Binding of 53BP1 to the Prb Tumor Suppressor.
Carr, S.M., Munro, S., Zalmas, L., Fedorov, O., Johansson, C., Krojer, T., Sagum, C.A., Bedford, M.T., Oppermann, U., La Thangue, N.B.(2014) Proc Natl Acad Sci U S A 111: 11341
- PubMed: 25049398
- DOI: https://doi.org/10.1073/pnas.1403737111
- Primary Citation of Related Structures:
4CRI - PubMed Abstract:
The retinoblastoma tumor suppressor protein pRb is a key regulator of cell cycle progression and mediator of the DNA damage response. Lysine methylation at K810, which occurs within a critical Cdk phosphorylation motif, holds pRb in the hypophosphorylated growth-suppressing state. We show here that methyl K810 is read by the tandem tudor domain containing tumor protein p53 binding protein 1 (53BP1). Structural elucidation of 53BP1 in complex with a methylated K810 pRb peptide emphasized the role of the 53BP1 tandem tudor domain in recognition of the methylated lysine and surrounding residues. Significantly, binding of 53BP1 to methyl K810 occurs on E2 promoter binding factor target genes and allows pRb activity to be effectively integrated with the DNA damage response. Our results widen the repertoire of cellular targets for 53BP1 and suggest a previously unidentified role for 53BP1 in regulating pRb tumor suppressor activity.
- Department of Oncology, University of Oxford, Headington, Oxford OX3 7DQ, United Kingdom;
Organizational Affiliation: