Aminoacetone Oxidase from Streptococcus Oligofermentas Belongs to a New Three-Domain Family of Bacterial Flavoproteins.
Molla, G., Nardini, M., Motta, P., D'Arrigo, P., Panzeri, W., Pollegioni, L.(2014) Biochem J 464: 387
- PubMed: 25269103
- DOI: https://doi.org/10.1042/BJ20140972
- Primary Citation of Related Structures:
4CNJ, 4CNK - PubMed Abstract:
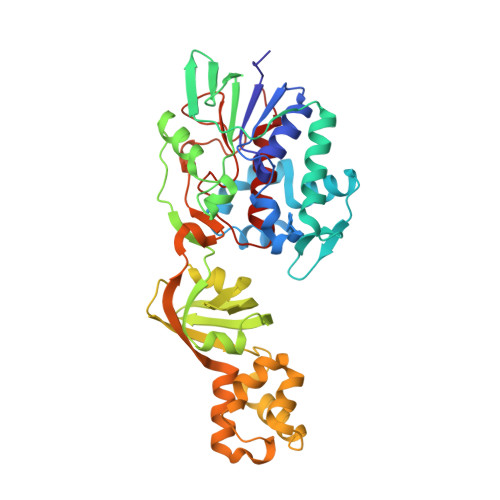
The aaoSo gene from Streptococcus oligofermentans encodes a 43 kDa flavoprotein, aminoacetone oxidase (SoAAO), which was reported to possess a low catalytic activity against several different L-amino acids; accordingly, it was classified as an L-amino acid oxidase. Subsequently, SoAAO was demonstrated to oxidize aminoacetone (a pro-oxidant metabolite), with an activity ~25-fold higher than the activity displayed on L-lysine, thus lending support to the assumption of aminoacetone as the preferred substrate. In the present study, we have characterized the SoAAO structure-function relationship. SoAAO is an FAD-containing enzyme that does not possess the classical properties of the oxidase/dehydrogenase class of flavoproteins (i.e. no flavin semiquinone formation is observed during anaerobic photoreduction as well as no reaction with sulfite) and does not show a true L-amino acid oxidase activity. From a structural point of view, SoAAO belongs to a novel protein family composed of three domains: an α/β domain corresponding to the FAD-binding domain, a β-domain partially modulating accessibility to the coenzyme, and an additional α-domain. Analysis of the reaction products of SoAAO on aminoacetone showed 2,5-dimethylpyrazine as the main product; we propose that condensation of two aminoacetone molecules yields 3,6-dimethyl-2,5-dihydropyrazine that is subsequently oxidized to 2,5-dimethylpyrazine. The ability of SoAAO to bind two molecules of the substrate analogue O-methylglycine ligand is thought to facilitate the condensation reaction. A specialized role for SoAAO in the microbial defence mechanism related to aminoacetone catabolism through a pathway yielding dimethylpyrazine derivatives instead of methylglyoxal can be proposed.
- *Dipartimento di Biotecnologie e Scienze della Vita, Università degli Studi deII'Insubria, via J.H. Dunant 3, 21100 Varese, ltaly.
Organizational Affiliation: