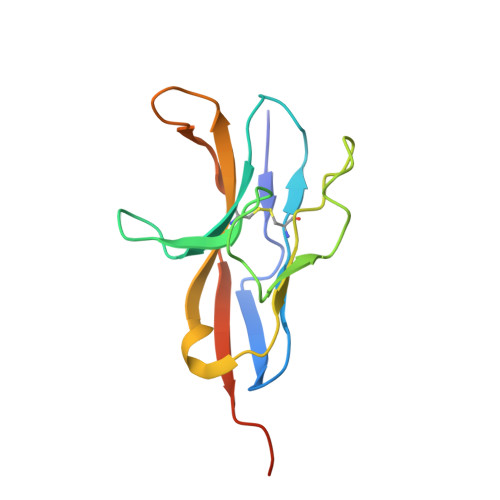
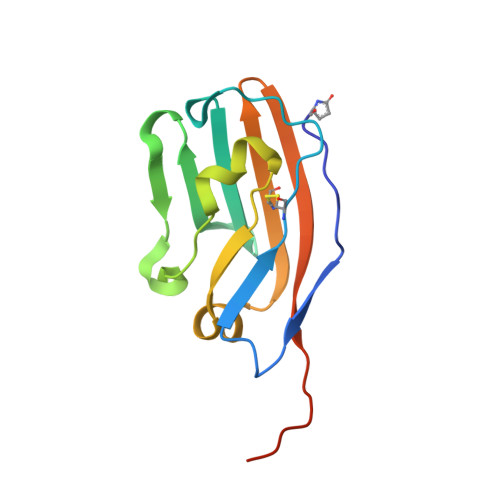
Polymorphisms in the Human Inhibitory Signal-Regulatory Protein Alpha Do not Affect Binding to its Ligand Cd47.
Hatherley, D., Lea, S.M., Johnson, S., Barclay, A.N.(2014) J Biological Chem 289: 10024
- PubMed: 24550402
- DOI: https://doi.org/10.1074/jbc.M114.550558
- Primary Citation of Related Structures:
4CMM - PubMed Abstract:
CD47 is a widely distributed membrane protein that interacts with signal-regulatory protein α (SIRPα), an inhibitory receptor on myeloid cells that gives a "don't-eat-me" signal. Manipulation of the interaction is of considerable interest in the immunotherapy of cancer and in xenotransplantation. The amino-terminal ligand binding domain of SIRPα is highly polymorphic in contrast to the single Ig-like domain of CD47. There is confusion as to whether the polymorphisms will affect ligand binding, but this is an important point for this interaction and other paired receptors being considered as targets for therapy. We show by x-ray crystallography that one human SIRPα allele differing in 13 amino acid residues has a very similar binding site and that several different alleles all bind CD47 with similar affinity as expected because the residues are mostly surface-exposed and distant from the binding site. A peptide from the binding site of CD47 has been reported to mimic the CD47 interaction with SIRPα, but we could find no binding. We discuss the possible pitfalls in determining the affinity of weak interactions and also speculate on how SIRPα polymorphisms may have been selected by pathogens and how this may also be true in other paired receptors such as the KIRs.
- From the Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, United Kingdom.
Organizational Affiliation: